circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000164535:-:7:6432836:6436533
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000164535:-:7:6432836:6436533 | ENSG00000164535 | MSTRG.29469.1 | - | 7 | 6432837 | 6436533 | 522 | GAACGAUUUGUAACCCUGGACCGCGGAAGUCUAUGUCUAAGCUGCUUUACAUCCGCCUGGCGCUGUUUUUUCCAGAGAUGGUCUGGGCCUCUCUGGGGGCUGCCUGGGUGGCAGAUGGUGUUCAGUGCGACAGGACAGUUGUAAACGGCAUCAUCGCAACCGUCGUGGUCAGUUGGAUCAUCAUCGCUGCCACAGUGGUUUCCAUUAUCAUUGUCUUUGACCCUCUUGGGGGGAAAAUGGCUCCAUAUUCCUCUGCCGGCCCCAGCCACCUGGAUAGUCAUGAUUCAAGCCAGUUACUUAAUGGCCUCAAGACAGCAGCUACAAGCGUGUGGGAAACCAGAAUCAAGCUCUUGUGCUGUUGCAUUGGGAAAGACGACCAUACUCGGGUUGCUUUUUCGAGACACAGAUCUGGUGCCCAGCGACAUUGCGGCGGGCCUCGCCCUGCUUCAUCAGCAACAGGACAAUAUCAGGAACAACCAAGAGCCUGCCCAGGUGGUCUGCCAUGCCCCAGGGAGCUCCCAGGAACGAUUUGUAACCCUGGACCGCGGAAGUCUAUGUCUAAGCUGCUUUAC | circ |
| ENSG00000164535:-:7:6432836:6436533 | ENSG00000164535 | MSTRG.29469.1 | - | 7 | 6432837 | 6436533 | 22 | GGAGCUCCCAGGAACGAUUUGU | bsj |
| ENSG00000164535:-:7:6432836:6436533 | ENSG00000164535 | MSTRG.29469.1 | - | 7 | 6436524 | 6436733 | 210 | ACUUCUUGUCACGUUUCCCAUCCUCAUUUUCUCACUUGCUGUCUGACUUAGGUGCUGAUUACCAAUUAUAAGUAGAAAAAGUCAGGAGGCGGGUGUGCACAGUACAAAUGUAUGCUCUGCAAAAGCUGUAUUUUUGCAAAAAGGAAGCUCUUCAUCUCUGAGGAAGCAACUGAGCAGUCGGAACGUUUUUUGUAUUUCAGGAACGAUUUG | ie_up |
| ENSG00000164535:-:7:6432836:6436533 | ENSG00000164535 | MSTRG.29469.1 | - | 7 | 6432637 | 6432846 | 210 | GAGCUCCCAGGUGAGCCUGGCUUCAUGGAGUGUCCAGUUCUGACACUUACACCUUUUGGUGGGUCUUCAGAAUCAUACAGCUAACCUAUAGCAAUUUCUUCCUCCCUUCCUUCAUUCCCCUUUCCCCUCCCUCCCCUCCCCCUCCUCCCCUUCCCUCCCCUCUCCCACCUUUUUUUUUUUUUUUUUUAAGACGGAGUCUCGCUCUGUCGC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
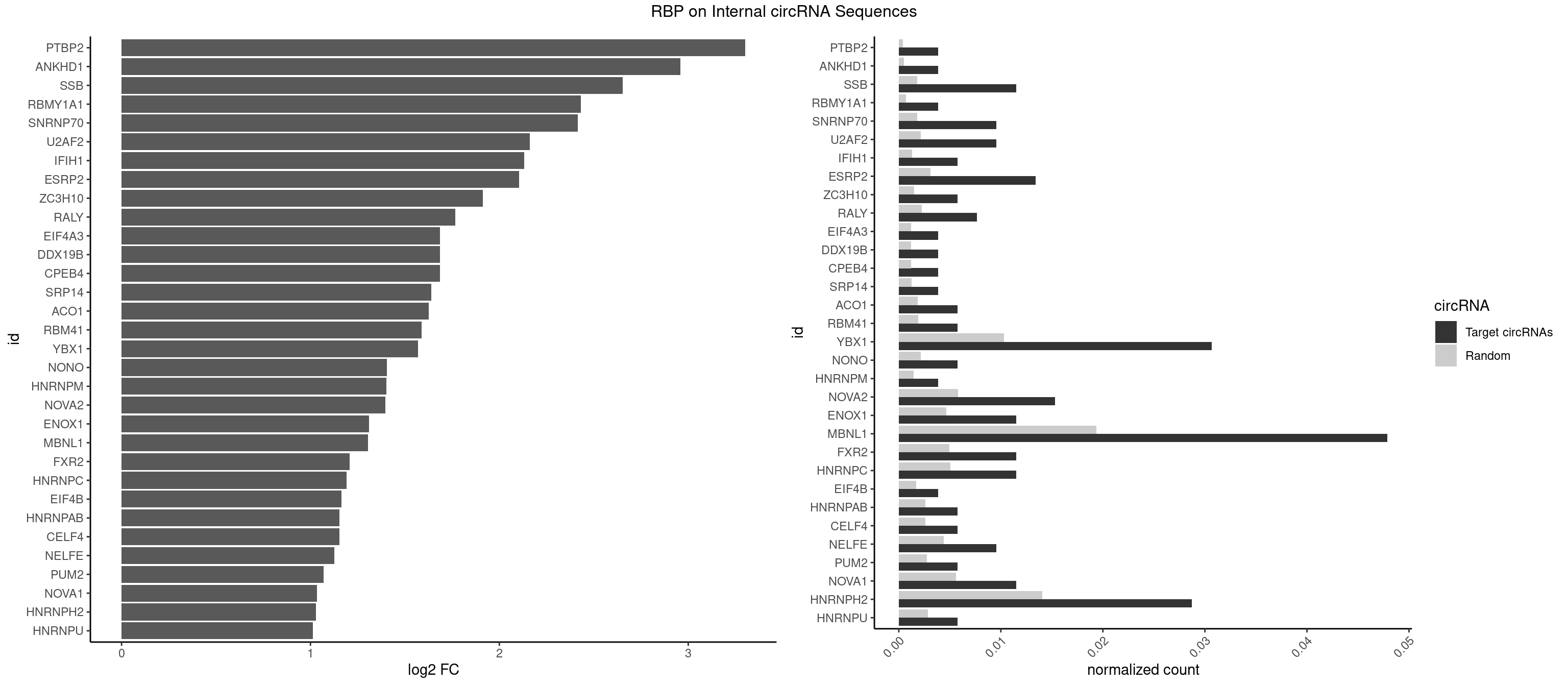
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 267 | 0.003831418 | 0.0003883867 | 3.302313 | CUCUCU | CUCUCU |
| ANKHD1 | 1 | 339 | 0.003831418 | 0.0004927293 | 2.959011 | AGACGA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SSB | 5 | 1260 | 0.011494253 | 0.0018274462 | 2.653012 | CUGUUU,GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBMY1A1 | 1 | 489 | 0.003831418 | 0.0007101099 | 2.431764 | CAAGAC | ACAAGA,CAAGAC |
| SNRNP70 | 4 | 1237 | 0.009578544 | 0.0017941145 | 2.416534 | AAUCAA,AUCAAG,AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| U2AF2 | 4 | 1477 | 0.009578544 | 0.0021419234 | 2.160899 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| IFIH1 | 2 | 904 | 0.005747126 | 0.0013115296 | 2.131590 | CCGCGG,CGCGGA | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ESRP2 | 6 | 2150 | 0.013409962 | 0.0031172377 | 2.104965 | GGGAAA,GGGGAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| ZC3H10 | 2 | 1053 | 0.005747126 | 0.0015274610 | 1.911705 | CAGCGA,CCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RALY | 3 | 1553 | 0.007662835 | 0.0022520629 | 1.766631 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| CPEB4 | 1 | 821 | 0.003831418 | 0.0011912456 | 1.685407 | UUUUUU | UUUUUU |
| DDX19B | 1 | 821 | 0.003831418 | 0.0011912456 | 1.685407 | UUUUUU | UUUUUU |
| EIF4A3 | 1 | 821 | 0.003831418 | 0.0011912456 | 1.685407 | UUUUUU | UUUUUU |
| SRP14 | 1 | 847 | 0.003831418 | 0.0012289250 | 1.640481 | CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ACO1 | 2 | 1283 | 0.005747126 | 0.0018607779 | 1.626935 | CAGUGC,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM41 | 2 | 1318 | 0.005747126 | 0.0019115000 | 1.588136 | UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| YBX1 | 15 | 7119 | 0.030651341 | 0.0103183321 | 1.570740 | AUCAUC,CAACCA,CAGCAA,CAUCAU,CAUCGC,CCCUGC,GAUCUG,GCCUGC,GGUCUG,GUCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| NONO | 2 | 1498 | 0.005747126 | 0.0021723567 | 1.403580 | AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| HNRNPM | 1 | 999 | 0.003831418 | 0.0014492040 | 1.402618 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| NOVA2 | 7 | 4013 | 0.015325670 | 0.0058171047 | 1.397577 | AGUCAU,AUCAUC,GAGACA,GGGGGG,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| ENOX1 | 5 | 3195 | 0.011494253 | 0.0046316558 | 1.311313 | AAGACA,AGACAG,AGGACA,GGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| MBNL1 | 24 | 13372 | 0.047892720 | 0.0193802045 | 1.305223 | AUGCCC,CCUGCU,CGCUGC,CGCUGU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCUU,CUUGUG,GCGCUG,GCUGCU,GCUUUU,GUGCUG,UCGCUG,UGCGGC,UGCUGU,UGCUUC,UGCUUU,UUGCUU,UUGUGC | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| FXR2 | 5 | 3434 | 0.011494253 | 0.0049780156 | 1.207270 | AGACAG,AGACGA,GACAGG,GGACAA,GGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPC | 5 | 3471 | 0.011494253 | 0.0050316361 | 1.191813 | CUUUUU,GGGGGG,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| EIF4B | 1 | 1179 | 0.003831418 | 0.0017100607 | 1.163831 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| CELF4 | 2 | 1782 | 0.005747126 | 0.0025839306 | 1.153273 | GGUGUU,GUGUGG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPAB | 2 | 1782 | 0.005747126 | 0.0025839306 | 1.153273 | AAAGAC,AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NELFE | 4 | 3028 | 0.009578544 | 0.0043896388 | 1.125704 | CUCUCU,CUCUGG,UCUCUG,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| PUM2 | 2 | 1890 | 0.005747126 | 0.0027404447 | 1.068431 | UACAUC,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| NOVA1 | 5 | 3872 | 0.011494253 | 0.0056127669 | 1.034129 | AUCAUC,GGGGGG,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| HNRNPH2 | 14 | 9711 | 0.028735632 | 0.0140746688 | 1.029740 | AUUGGG,CAGGAC,CUGGGG,GGGCUG,GGGGAA,GGGGCU,GGGGGA,GGGGGC,GGGGGG,UGGGGG,UGGGUG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPU | 2 | 1965 | 0.005747126 | 0.0028491350 | 1.012317 | GGGGGG,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
RBP on BSJ (Exon Only)
Plot
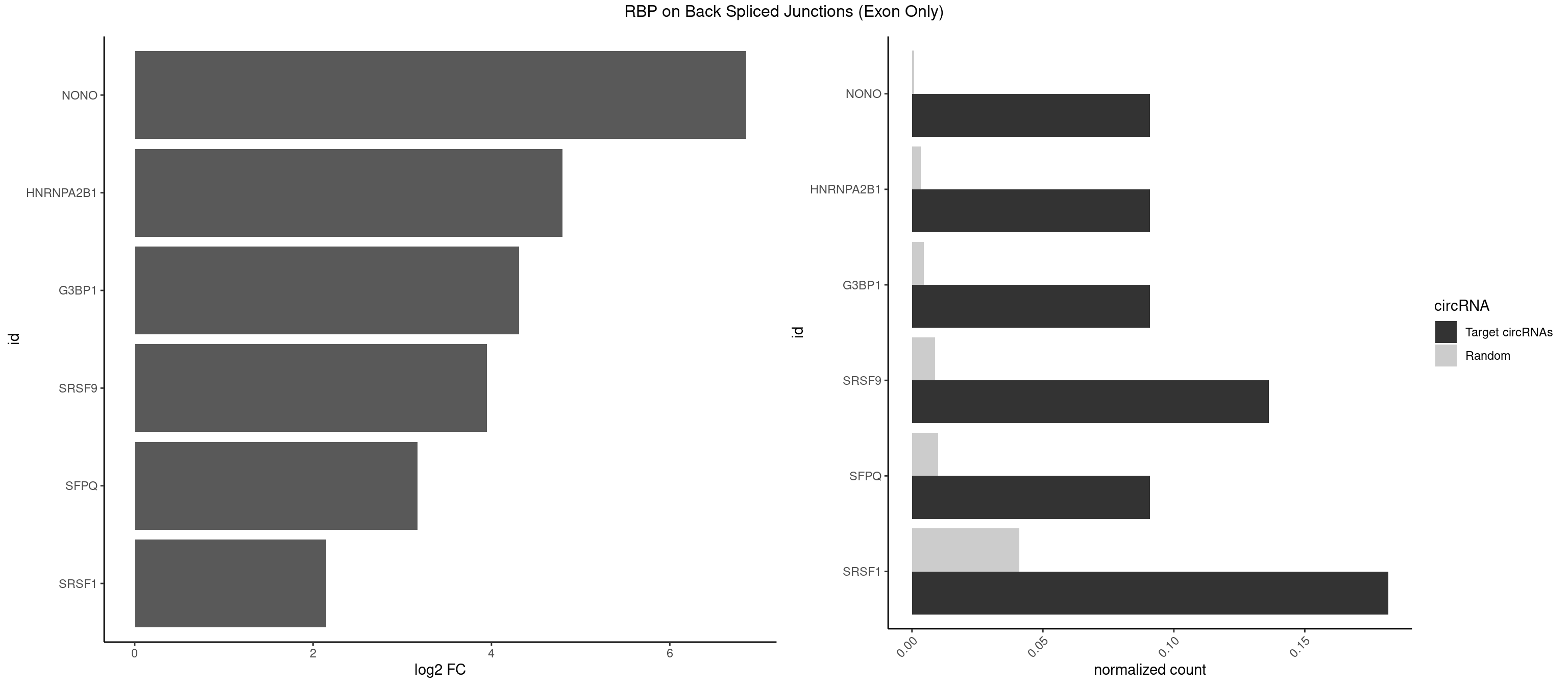
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NONO | 1 | 11 | 0.09090909 | 0.0007859576 | 6.853829 | AGGAAC | AGAGGA,AGGAAC,GAGGAA |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.0032748232 | 4.794936 | AGGAAC | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| G3BP1 | 1 | 69 | 0.09090909 | 0.0045847524 | 4.309509 | CCCAGG | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| SRSF9 | 2 | 134 | 0.13636364 | 0.0088420225 | 3.946939 | AGGAAC,GGAACG | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | AGGAAC | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF1 | 3 | 625 | 0.18181818 | 0.0410007860 | 2.148773 | AGGAAC,CCAGGA,CCCAGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
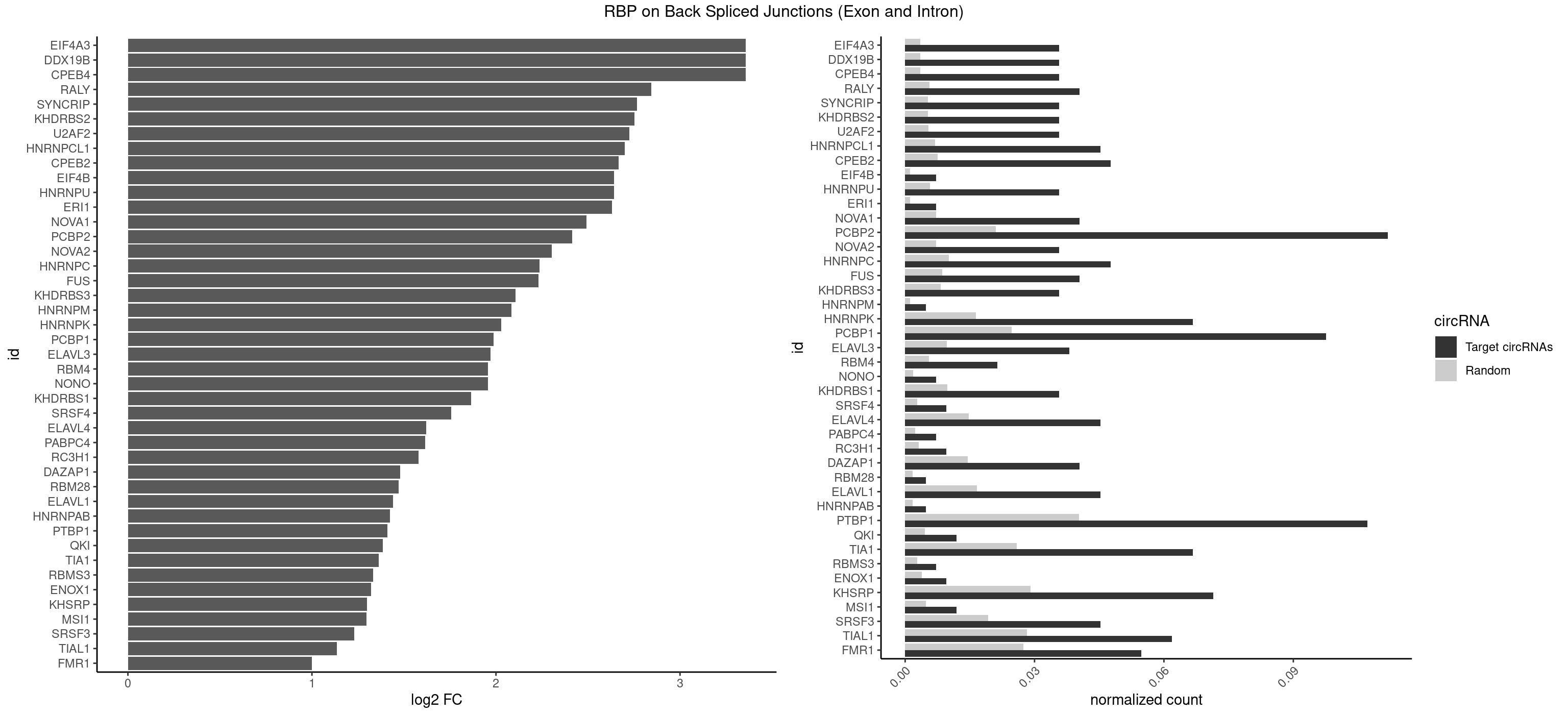
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CPEB4 | 14 | 691 | 0.035714286 | 0.003486497 | 3.356651 | UUUUUU | UUUUUU |
| DDX19B | 14 | 691 | 0.035714286 | 0.003486497 | 3.356651 | UUUUUU | UUUUUU |
| EIF4A3 | 14 | 691 | 0.035714286 | 0.003486497 | 3.356651 | UUUUUU | UUUUUU |
| RALY | 16 | 1117 | 0.040476190 | 0.005632809 | 2.845147 | UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| SYNCRIP | 14 | 1041 | 0.035714286 | 0.005249899 | 2.766140 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 14 | 1051 | 0.035714286 | 0.005300282 | 2.752360 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| U2AF2 | 14 | 1071 | 0.035714286 | 0.005401048 | 2.725190 | UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| HNRNPCL1 | 18 | 1381 | 0.045238095 | 0.006962918 | 2.699774 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB2 | 19 | 1487 | 0.047619048 | 0.007496977 | 2.667158 | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| EIF4B | 2 | 226 | 0.007142857 | 0.001143692 | 2.642803 | GUCGGA,UCGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPU | 14 | 1136 | 0.035714286 | 0.005728537 | 2.640263 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| ERI1 | 2 | 228 | 0.007142857 | 0.001153769 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| NOVA1 | 16 | 1428 | 0.040476190 | 0.007199718 | 2.491061 | UCAUUC,UUCAUU,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| PCBP2 | 46 | 4164 | 0.111904762 | 0.020984482 | 2.414877 | AUUACC,AUUCCC,CAAUUA,CCCCCU,CCCUCC,CCCUUC,CCUCCC,CCUCCU,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUCCC,CUUCCU,UUUUUU | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| NOVA2 | 14 | 1434 | 0.035714286 | 0.007229948 | 2.304444 | UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| HNRNPC | 19 | 2006 | 0.047619048 | 0.010111850 | 2.235492 | AUUUUU,CUUUUU,UUUUUA,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| FUS | 16 | 1711 | 0.040476190 | 0.008625554 | 2.230384 | GGGUGU,UGGUGG,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| KHDRBS3 | 14 | 1646 | 0.035714286 | 0.008298065 | 2.105654 | UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.001123539 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| HNRNPK | 27 | 3244 | 0.066666667 | 0.016349254 | 2.027741 | CCAUCC,CCCCCU,CCCCUU,UCCCAC,UCCCAU,UCCCCC,UCCCCU,UCCCUU,UUUUUU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| PCBP1 | 40 | 4891 | 0.097619048 | 0.024647320 | 1.985732 | AUUACC,AUUCCC,CAAUUA,CAUUCC,CCAUCC,CCCACC,CCCUCC,CCCUCU,CCCUUC,CCUCCC,CCUUCC,CCUUUC,CUAACC,CUUCCC,CUUUCC,UUUUUU | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| ELAVL3 | 15 | 1928 | 0.038095238 | 0.009718863 | 1.970751 | UAUUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| RBM4 | 8 | 1094 | 0.021428571 | 0.005516929 | 1.957598 | CCUUCC,CUUCCU,CUUCUU,UCCUUC,UUCCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| NONO | 2 | 364 | 0.007142857 | 0.001838976 | 1.957598 | AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| KHDRBS1 | 14 | 1945 | 0.035714286 | 0.009804514 | 1.864983 | UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| SRSF4 | 3 | 558 | 0.009523810 | 0.002816405 | 1.757684 | AGGAAG,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ELAVL4 | 18 | 2916 | 0.045238095 | 0.014696695 | 1.622046 | UAUUUU,UUGUAU,UUUGUA,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.002332729 | 1.614483 | AAAAAG | AAAAAA,AAAAAG |
| RC3H1 | 3 | 631 | 0.009523810 | 0.003184200 | 1.580608 | CCCUUC,UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| DAZAP1 | 16 | 2878 | 0.040476190 | 0.014505240 | 1.480499 | AGGAAG,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.001718057 | 1.470761 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ELAVL1 | 18 | 3309 | 0.045238095 | 0.016676743 | 1.439701 | UAUUUU,UUUGGU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.001773478 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PTBP1 | 44 | 8003 | 0.107142857 | 0.040326481 | 1.409736 | AGCUGU,AUUUUC,AUUUUU,CCUUCC,CCUUUC,CCUUUU,CUCCCC,CUCUUC,CUUCCU,CUUCUU,CUUUCC,CUUUUU,GCUCCC,UAUUUU,UCCCCC,UUCCCC,UUCCUU,UUCUUC,UUCUUG,UUUCCC,UUUCUU,UUUUCU,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| QKI | 4 | 904 | 0.011904762 | 0.004559653 | 1.384543 | AAUCAU,AUCAUA,CUAACC,UAACCU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| TIA1 | 27 | 5140 | 0.066666667 | 0.025901854 | 1.363910 | AUUUUC,AUUUUU,CCUUUC,CUUUUG,CUUUUU,GUUUUU,UAUUUU,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBMS3 | 2 | 562 | 0.007142857 | 0.002836558 | 1.332360 | CUAUAG,UAUAGC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ENOX1 | 3 | 756 | 0.009523810 | 0.003813986 | 1.320239 | AGUACA,AUACAG,CAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| KHSRP | 29 | 5764 | 0.071428571 | 0.029045748 | 1.298174 | AUGUAU,CACCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCUUCC,CUCCCU,CUGUAU,GGCUUC,UAUUUU,UCCCUC,UCCUUC,UGUAUG,UUGUAU | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| MSI1 | 4 | 961 | 0.011904762 | 0.004846836 | 1.296424 | AGGAAG,AGGAGG,UAGGUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| SRSF3 | 18 | 3825 | 0.045238095 | 0.019276501 | 1.230695 | AACGAU,ACGAUU,ACUUCU,CUCUUC,CUUCAG,CUUCAU,CUUCUU,GCUUCA,UACAAA,UACAGC,UCAUCU,UCUUCA,UCUUCC,UUCAUC,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| TIAL1 | 25 | 5597 | 0.061904762 | 0.028204353 | 1.134133 | AUUUUC,AUUUUU,CUUUUG,CUUUUU,GUUUUU,UAUUUU,UUUCAG,UUUUAA,UUUUUA,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| FMR1 | 22 | 5430 | 0.054761905 | 0.027362958 | 1.000948 | AAGGAA,AGGAAG,AGGAGG,AUGGAG,CUGAGG,UGAGGA,UGGAGU,UUUUUU | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.