circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000219481:-:1:16579192:16583696
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000219481:-:1:16579192:16583696 | ENSG00000219481 | ENST00000430580 | - | 1 | 16579193 | 16583696 | 813 | AAUAUGAAGAGUGCAAAGACCUCAUAAAAUCUAUGCUGAGGAAUGAGCGACAGUUCAAGGAGGAGAAGCUUGCAGAGCAGCUCAAGCAAGCUGAGGAGCUCAGGCAAUAUAAAGUCCUGGUUCACUCUCAGGAACGAGAGCUGACCCAGUUAAGGGAGAAGUUACGGGAAGGGAGAGAUGCCUCCCGCUCAUUGAAUCAGCAUCUCCAGGCCCUCCUCACUCCGGAUAAGCCAGACAAGUCCCAGGGGCAGGACCUCCAAGAACAGCUGGCUGAGGGGUGUAGACUGGCACAGCAACUUUUCCAGAAGCUCAGCCCAGAAAAUGACGAAGAUGAGGAUGAAGAUGUUCAAGUUGAGGAGGCUGAGAAAGUACUGGAAUCAUCUGCCCCCAGGGAGGUGCAGAAGGCUGAAGAAAGCAAAGUCCCUGAGGACUCACUGGAGGAAUGUGCCAUCACUUGUUCAAAUAGCCACGGCCCUUGUGACUCCAACCAGCCUCACAAGAACAUCAACAUCACCUUUGAGGAAGACAAAGUCAACUCAGCUCUGGUUGUAGACAGAGAAUCCUCUCAUGAUGAAUGUCAGGAUGCUGUAAACAUUCUCCCAGUCCCUGGCCCCACCUCUUCUGCCACAAACGUCAGCAUGGUGGUAUCAGCCGGCCCUUUGUCCAGCGAGAAGGCAGAGAUGAACAUUCUAGAAAUCAACGAGAAAUUGCAUCCCCAGCUGGCAGAGAAGAAACAGCAGUUCAGAAACCUCAAAGAGAAAUGUUUUGUAACUCAACUGGCCUGCUUCCUGGCCAACCAGCAGAACAAAUACAAAUAUGAAGAGUGCAAAGACCUCAUAAAAUCUAUGCUGAGGAAUGAGCGA | circ |
| ENSG00000219481:-:1:16579192:16583696 | ENSG00000219481 | ENST00000430580 | - | 1 | 16579193 | 16583696 | 22 | GAACAAAUACAAAUAUGAAGAG | bsj |
| ENSG00000219481:-:1:16579192:16583696 | ENSG00000219481 | ENST00000430580 | - | 1 | 16583687 | 16583896 | 210 | CUUUCUUCAUCUCUAAAUUUUGGAGGAUCAGAUGCCAGAAAGUCAGGAGACUGAAGAGUAAAGAUGUGGAAAUCCCUGUCUAGACCCUGGUACUGGGGAGAGUUUUGUCCUUGGGAUGGACCUGGCUCCUGCCCUGUAGGCAAUGACCACAGCAGCAUGUCCAGCCUUCCACUGAGGCAGGCGUGUCUGUCUUUUCUCAGAAUAUGAAGA | ie_up |
| ENSG00000219481:-:1:16579192:16583696 | ENSG00000219481 | ENST00000430580 | - | 1 | 16578993 | 16579202 | 210 | AACAAAUACAGUAAGAUCUAUAGGCUCACCAUCACGAAAGUGAUGAACGAAGUCCUGUCUUCUCUCUGAGAAACUAAGUGCUCUCUCCAUCUAAAAUAAUGUCAUCCUCCCCAUACUUCUAGGAAAACAGAAAUGGGUAUUUUAACAUUUUGUUAAAGUUGGAAGACAGAGGUCCCAAAAUAUUUAGCAACUUUCCAUGUUUGCAAUCAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
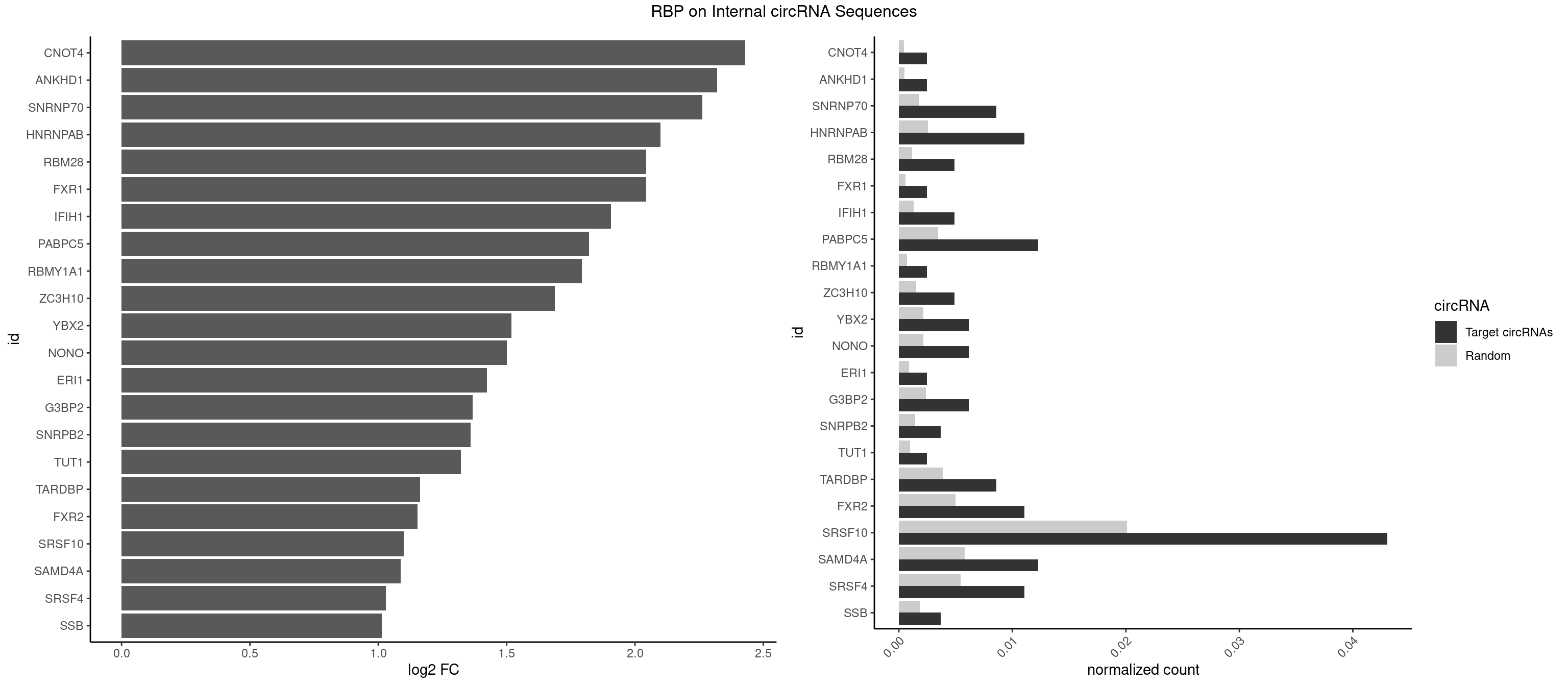
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 314 | 0.002460025 | 0.0004564992 | 2.429988 | GACAGA | GACAGA |
| ANKHD1 | 1 | 339 | 0.002460025 | 0.0004927293 | 2.319805 | GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SNRNP70 | 6 | 1237 | 0.008610086 | 0.0017941145 | 2.262756 | AAUCAA,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPAB | 8 | 1782 | 0.011070111 | 0.0025839306 | 2.099030 | AAAGAC,AAGACA,ACAAAG,AGACAA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM28 | 3 | 822 | 0.004920049 | 0.0011926949 | 2.044448 | GUGUAG,UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| FXR1 | 1 | 411 | 0.002460025 | 0.0005970720 | 2.042696 | AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| IFIH1 | 3 | 904 | 0.004920049 | 0.0013115296 | 1.907422 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| PABPC5 | 9 | 2400 | 0.012300123 | 0.0034795387 | 1.821705 | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBMY1A1 | 1 | 489 | 0.002460025 | 0.0007101099 | 1.792558 | ACAAGA | ACAAGA,CAAGAC |
| ZC3H10 | 3 | 1053 | 0.004920049 | 0.0015274610 | 1.687537 | CAGCGA,CCAGCG,GAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| YBX2 | 4 | 1480 | 0.006150062 | 0.0021462711 | 1.518769 | AACAUC,ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NONO | 4 | 1498 | 0.006150062 | 0.0021723567 | 1.501340 | AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ERI1 | 1 | 632 | 0.002460025 | 0.0009173461 | 1.423135 | UUCAGA | UUCAGA,UUUCAG |
| G3BP2 | 4 | 1644 | 0.006150062 | 0.0023839405 | 1.367253 | AGGAUG,GGAUAA,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SNRPB2 | 2 | 991 | 0.003690037 | 0.0014376103 | 1.359963 | AUUGCA,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| TUT1 | 1 | 678 | 0.002460025 | 0.0009840095 | 1.321929 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| TARDBP | 6 | 2654 | 0.008610086 | 0.0038476365 | 1.162055 | GAAUGA,GAAUGU,GUUUUG,UGAAUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| FXR2 | 8 | 3434 | 0.011070111 | 0.0049780156 | 1.153027 | AGACAA,AGACAG,GACAAA,GACAAG,GACAGA,GACGAA,UGACGA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| SRSF10 | 34 | 13860 | 0.043050431 | 0.0200874160 | 1.099736 | AAAGAC,AAAGAG,AAGAAA,AAGACA,AAGAGA,AAGGAG,AAGGGA,ACAAAG,AGACAA,AGAGAA,CAAAGA,GACAAA,GAGAAA,GAGAAG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SAMD4A | 9 | 3992 | 0.012300123 | 0.0057866714 | 1.087867 | CGGGAA,CUGGAA,CUGGCA,CUGGCC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SRSF4 | 8 | 3740 | 0.011070111 | 0.0054214720 | 1.029913 | AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| SSB | 2 | 1260 | 0.003690037 | 0.0018274462 | 1.013806 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
RBP on BSJ (Exon Only)
Plot
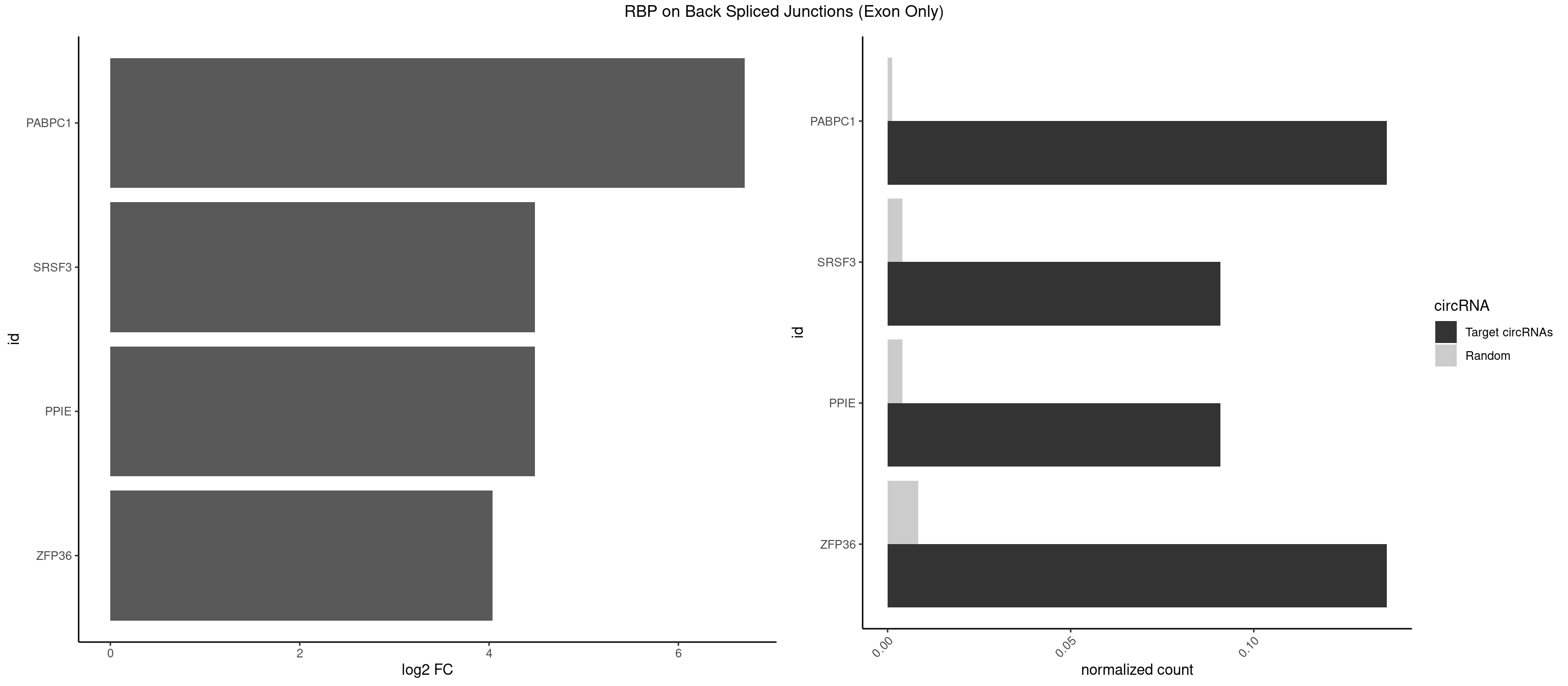
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PABPC1 | 2 | 19 | 0.13636364 | 0.001309929 | 6.701826 | ACAAAU,CAAAUA | AAAAAA,ACAAAC,ACGAAU,AGAAAA,CAAACC,CAAAUA,CGAACA,CUAAUA,GAAAAA,GAAAAC |
| PPIE | 1 | 61 | 0.09090909 | 0.004060781 | 4.484596 | AAAUAU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAU,AUAAUA,AUAUAA,AUAUAU,AUAUUU,AUUAAA,AUUUAA,UAAAAA,UAAAUA,UAAAUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUU,UUAAAA,UUAAAU,UUAUUA |
| SRSF3 | 1 | 61 | 0.09090909 | 0.004060781 | 4.484596 | UACAAA | AACGAU,ACCACC,ACUACG,AGAGAU,CACAAC,CACAUC,CACCAC,CAGAGA,CAUCAC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CUACAG,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUUAU,GACUUC,GAGAUU,UACAGC,UCAGAG,UCUCCA,UCUUCA,UCUUCC,UGUCAA,UUCGAC,UUCUCC,UUCUUC |
| ZFP36 | 2 | 126 | 0.13636364 | 0.008318051 | 4.035070 | ACAAAU,CAAAUA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
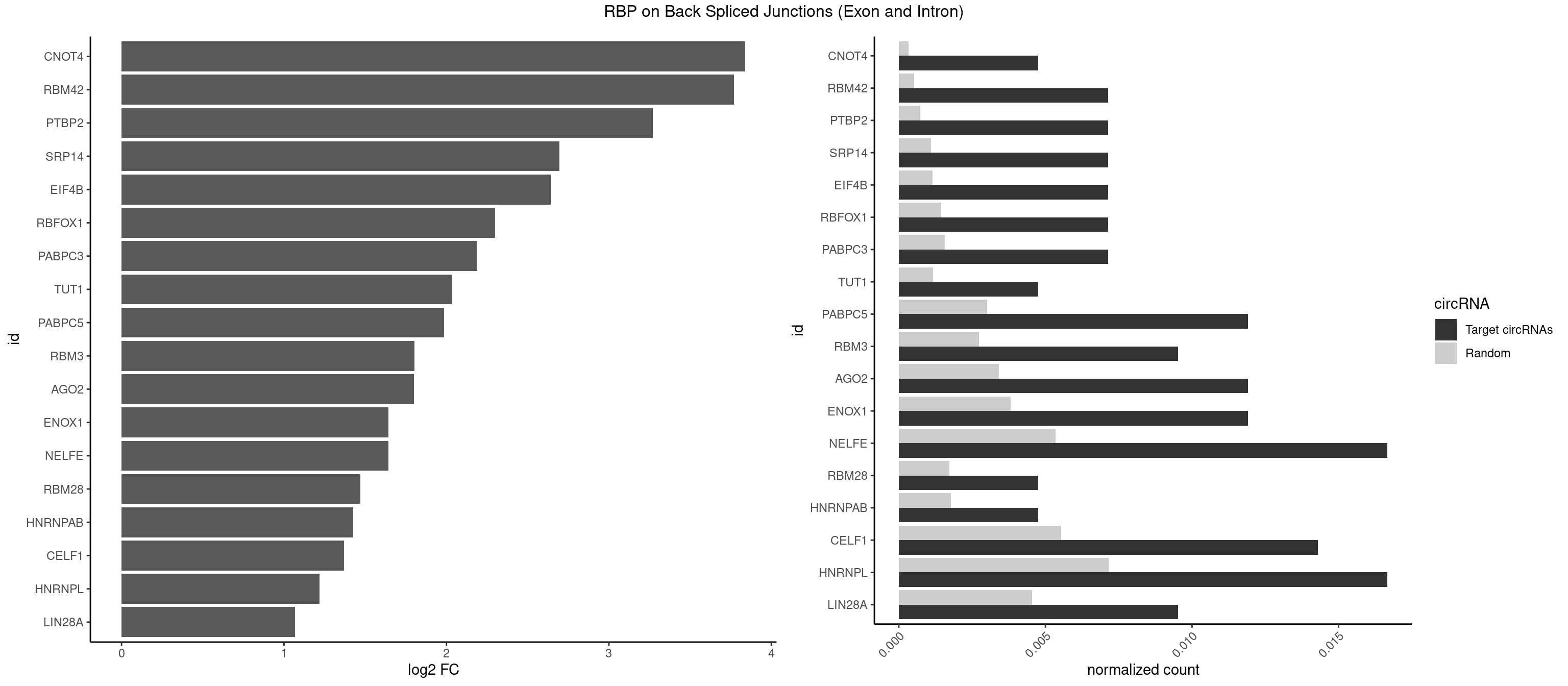
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| RBM42 | 2 | 103 | 0.007142857 | 0.0005239823 | 3.768911 | AACUAA,ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| PTBP2 | 2 | 146 | 0.007142857 | 0.0007406288 | 3.269679 | CUCUCU | CUCUCU |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CCUGUA,CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | GUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | AGCAUG,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC5 | 4 | 596 | 0.011904762 | 0.0030078597 | 1.984730 | AGAAAG,AGAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAACUA,GAAACU,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| AGO2 | 4 | 677 | 0.011904762 | 0.0034159613 | 1.801175 | AAAGUG,AAGUGC,AGUGCU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| ENOX1 | 4 | 756 | 0.011904762 | 0.0038139863 | 1.642167 | AAGACA,AAUACA,AGACAG,AUACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| NELFE | 6 | 1059 | 0.016666667 | 0.0053405885 | 1.641895 | CUCUCU,CUGGCU,UCUCUC,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CELF1 | 5 | 1097 | 0.014285714 | 0.0055320435 | 1.368689 | CUGUCU,UGUCUG,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPL | 6 | 1419 | 0.016666667 | 0.0071543732 | 1.220068 | AAAUAA,AAAUAC,AACAAA,AAUACA,ACCACA,CACGAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | AGGAGA,GGAGGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.