circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000145907:+:5:151786571:151803996
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000145907:+:5:151786571:151803996 | ENSG00000145907 | MSTRG.27198.12 | + | 5 | 151786572 | 151803996 | 1355 | GUUUGGACAUAUUUGACUCUUUUCCCCCCAGGUUGAAUUGACCAAAGCAAUGGUGAUGGAGAAGCCUAGUCCCCUGCUGGUCGGGCGGGAAUUUGUGAGACAGUAUUACACACUGCUGAACCAGGCCCCAGACAUGCUGCAUAGAUUUUAUGGAAAGAACUCUUCUUAUGUCCAUGGGGGAUUGGAUUCAAAUGGAAAGCCAGCAGAUGCAGUCUACGGACAGAAAGAAAUCCACAGGAAAGUGAUGUCACAAAACUUCACCAACUGCCACACCAAGAUUCGCCAUGUUGAUGCUCAUGCCACGCUAAAUGAUGGUGUGGUAGUCCAGGUGAUGGGGCUUCUCUCUAACAACAACCAGGCUUUGAGGAGAUUCAUGCAAACGUUUGUCCUUGCUCCUGAGGGGUCUGUUGCAAAUAAAUUCUAUGUUCACAAUGAUAUCUUCAGAUACCAAGAUGAGGUCUUUGGUGGGUUUGUCACUGAGCCUCAGGAGGAGUCUGAAGAAGAAGUAGAGGAACCUGAAGAAAGACAGCAAACACCUGAGGUGGUACCUGAUGAUUCUGGAACUUUCUAUGAUCAGGCAGUUGUCAGUAAUGACAUGGAAGAACAUUUAGAGGAGCCUGUUGCUGAACCAGAGCCUGAUCCUGAACCAGAACCAGAACAAGAACCUGUAUCUGAAAUCCAAGAGGAAAAGCCUGAGCCAGUAUUAGAAGAAACUGCCCCUGAGGAUGCUCAGAAGAGUUCUUCUCCAGCACCUGCAGACAUAGCUCAGACAGUACAGGAAGACUUGAGGACAUUUUCUUGGGCAUCUGUGACCAGUAAGAAUCUUCCACCCAGUGGAGCUGUUCCAGUUACUGGGAUACCACCUCAUGUUGUUAAAGUACCAGCUUCACAGCCCCGUCCAGAGUCUAAGCCUGAAUCUCAGAUUCCACCACAAAGACCUCAGCGGGAUCAAAGAGUGCGAGAACAACGAAUAAAUAUUCCUCCCCAAAGGGGACCCAGACCAAUCCGUGAGGCUGGUGAGCAAGGUGACAUUGAACCCCGAAGAAUGGUGAGACACCCUGACAGUCACCAACUCUUCAUUGGCAACCUGCCUCAUGAAGUGGACAAAUCAGAGCUUAAAGAUUUCUUUCAAAGUUAUGGAAACGUGGUGGAGUUGCGCAUUAACAGUGGUGGGAAAUUACCCAAUUUUGGUUUUGUUGUGUUUGAUGAUUCUGAGCCUGUUCAGAAAGUCCUUAGCAACAGGCCCAUCAUGUUCAGAGGUGAGGUCCGUCUGAAUGUCGAAGAGAAGAAGACUCGAGCUGCCAGGGAAGGCGACCGACGAGAUAAUCGCCUUCGGGGACCUGGAGGCCCUCGAGGUUUGGACAUAUUUGACUCUUUUCCCCCCAGGUUGAAUUGACCAAAGCAA | circ |
| ENSG00000145907:+:5:151786571:151803996 | ENSG00000145907 | MSTRG.27198.12 | + | 5 | 151786572 | 151803996 | 22 | AGGCCCUCGAGGUUUGGACAUA | bsj |
| ENSG00000145907:+:5:151786571:151803996 | ENSG00000145907 | MSTRG.27198.12 | + | 5 | 151786372 | 151786581 | 210 | GAUGAAUUCUUCAUAAUGAUUUCCCCCUUUUGUAUUCAUAUGAUUGCAAUACAUUUGUAUAAAAUCUCAACUGGUCUGUAUGGGCUCUGUUUGUUUUUAUGAUGUUUGUUCAUGUUUUAAACGACUUGCUGAAAUGAUCUUGUCUCUGAGUUGGUUUCAGCUAAAUGAUUCGGUCUUUUCCCCUGUGUUUGAUCCUUCAGGUUUGGACAU | ie_up |
| ENSG00000145907:+:5:151786571:151803996 | ENSG00000145907 | MSTRG.27198.12 | + | 5 | 151803987 | 151804196 | 210 | GGCCCUCGAGGUGGGCUGGGUGGUGGAAUGAGAGGCCCUCCCCGUGGAGGCAUGGUGCAGAAACCAGGAUUUGGAGUGGGAAGGGGGCUUGCGCCACGGCAGUGAAUCUUCAUGGAUCUUCAUGCAGCCAUACAAACCCUGGUUCCAACAGAAUGGUGAAUUUUCGACAGCCUUUGGUAUCUUGGAGUAUGACCCCAGUCUGUUAUAAAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
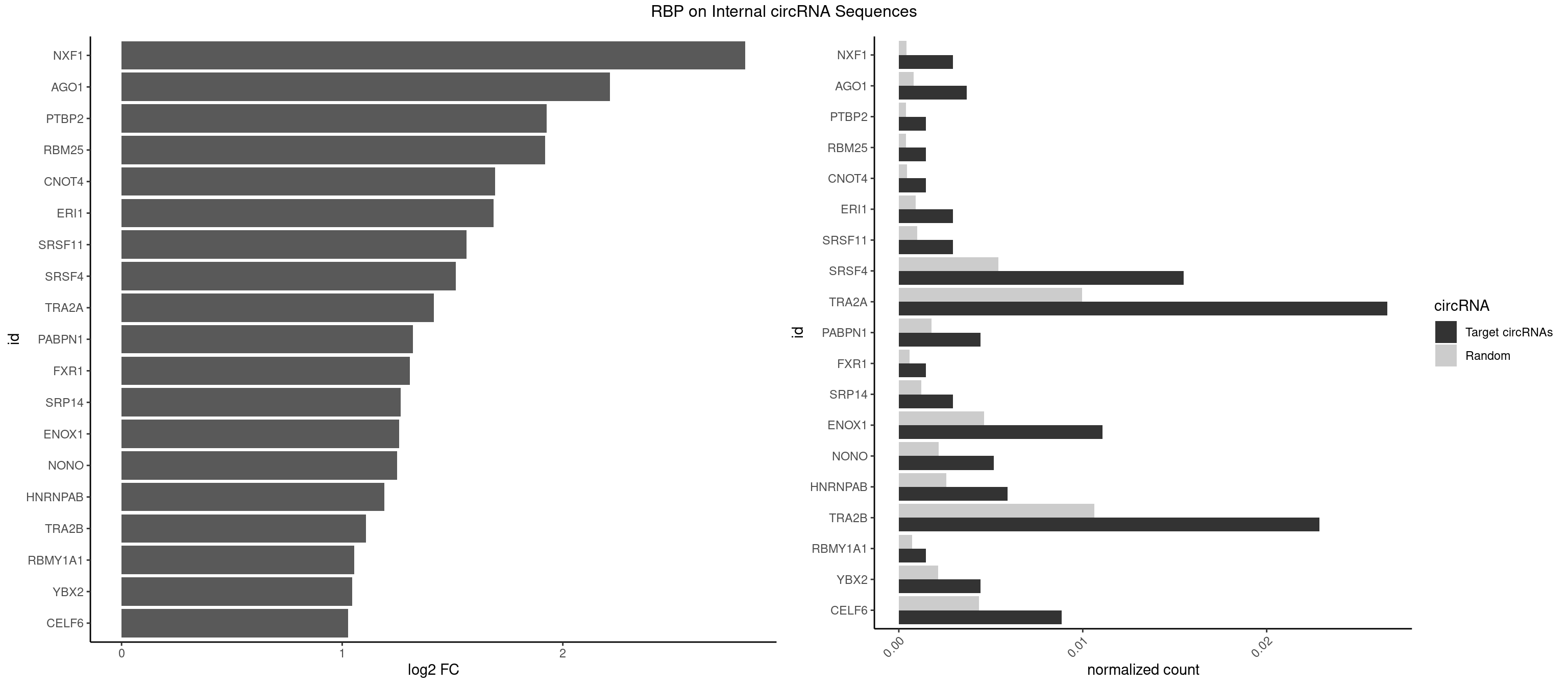
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 3 | 286 | 0.002952030 | 0.0004159215 | 2.827324 | AACCUG | AACCUG |
| AGO1 | 4 | 548 | 0.003690037 | 0.0007956130 | 2.213497 | GGUAGU,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| PTBP2 | 1 | 267 | 0.001476015 | 0.0003883867 | 1.926142 | CUCUCU | CUCUCU |
| RBM25 | 1 | 268 | 0.001476015 | 0.0003898359 | 1.920768 | UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| CNOT4 | 1 | 314 | 0.001476015 | 0.0004564992 | 1.693023 | GACAGA | GACAGA |
| ERI1 | 3 | 632 | 0.002952030 | 0.0009173461 | 1.686169 | UUCAGA | UUCAGA,UUUCAG |
| SRSF11 | 3 | 688 | 0.002952030 | 0.0009985015 | 1.563871 | AAGAAG | AAGAAG |
| SRSF4 | 20 | 3740 | 0.015498155 | 0.0054214720 | 1.515340 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| TRA2A | 35 | 6871 | 0.026568266 | 0.0099589296 | 1.415641 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| PABPN1 | 5 | 1222 | 0.004428044 | 0.0017723764 | 1.320985 | AGAAGA | AAAAGA,AGAAGA |
| FXR1 | 1 | 411 | 0.001476015 | 0.0005970720 | 1.305730 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRP14 | 3 | 847 | 0.002952030 | 0.0012289250 | 1.264310 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ENOX1 | 14 | 3195 | 0.011070111 | 0.0046316558 | 1.257070 | AAGACA,AGACAG,AGGACA,AGUACA,CAGACA,CGGACA,GGACAG,GUACAG,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| NONO | 6 | 1498 | 0.005166052 | 0.0021723567 | 1.249801 | AGAGGA,AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| HNRNPAB | 7 | 1782 | 0.005904059 | 0.0025839306 | 1.192140 | AAAGAC,AAGACA,ACAAAG,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| TRA2B | 30 | 7329 | 0.022878229 | 0.0106226650 | 1.106830 | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AGAAGA,AGGAAA,AGGAAG,GAAAGA,GAAGAA,GGAAGG,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBMY1A1 | 1 | 489 | 0.001476015 | 0.0007101099 | 1.055593 | ACAAGA | ACAAGA,CAAGAC |
| YBX2 | 5 | 1480 | 0.004428044 | 0.0021462711 | 1.044837 | AACAAC,ACAACA,ACAACG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF6 | 11 | 2997 | 0.008856089 | 0.0043447134 | 1.027409 | GUGAGG,GUGAUG,GUGGUG,GUGUGG,UGUGAG,UGUGGU,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
RBP on BSJ (Exon Only)
Plot
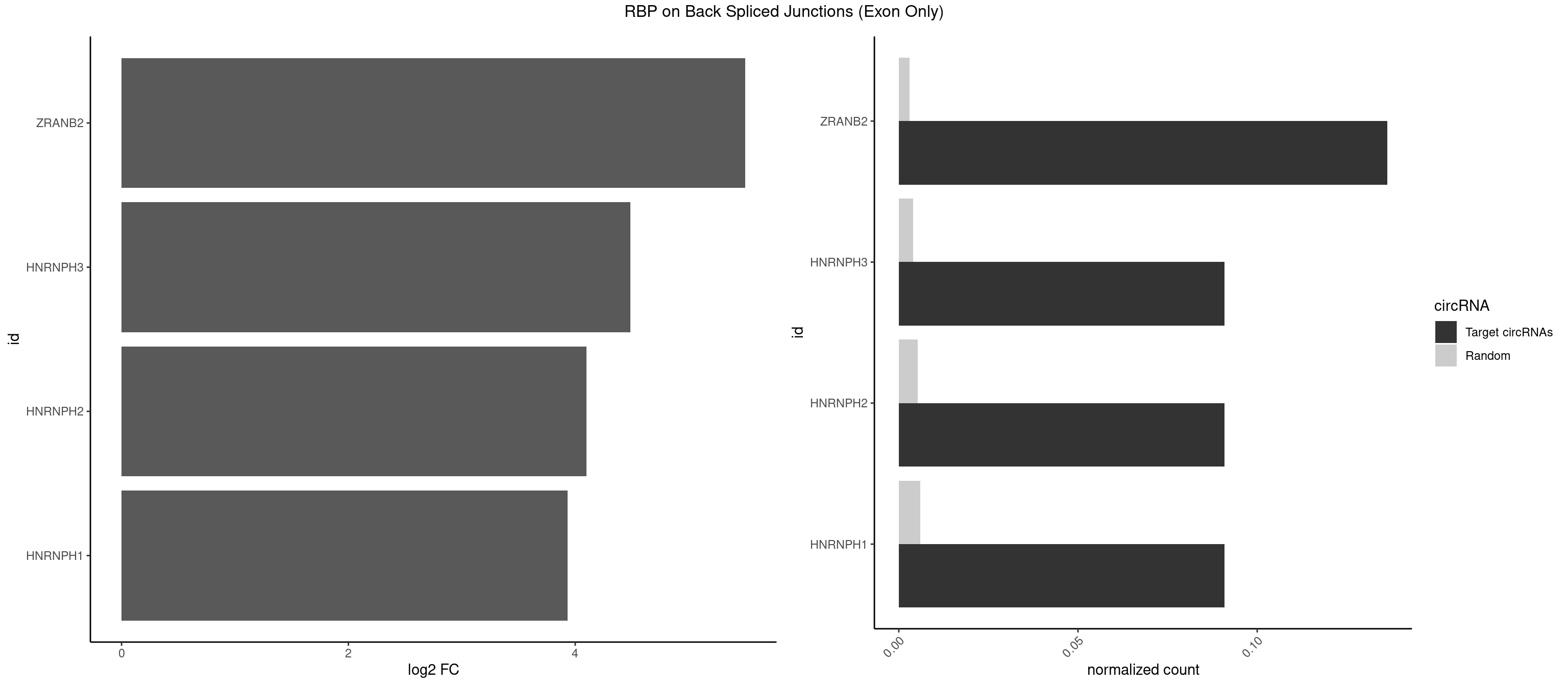
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZRANB2 | 2 | 45 | 0.13636364 | 0.003012837 | 5.500192 | AGGUUU,GAGGUU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| HNRNPH3 | 1 | 61 | 0.09090909 | 0.004060781 | 4.484596 | UCGAGG | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| HNRNPH2 | 1 | 80 | 0.09090909 | 0.005305214 | 4.098942 | UCGAGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPH1 | 1 | 90 | 0.09090909 | 0.005960178 | 3.930997 | UCGAGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
RBP on BSJ (Exon and Intron)
Plot
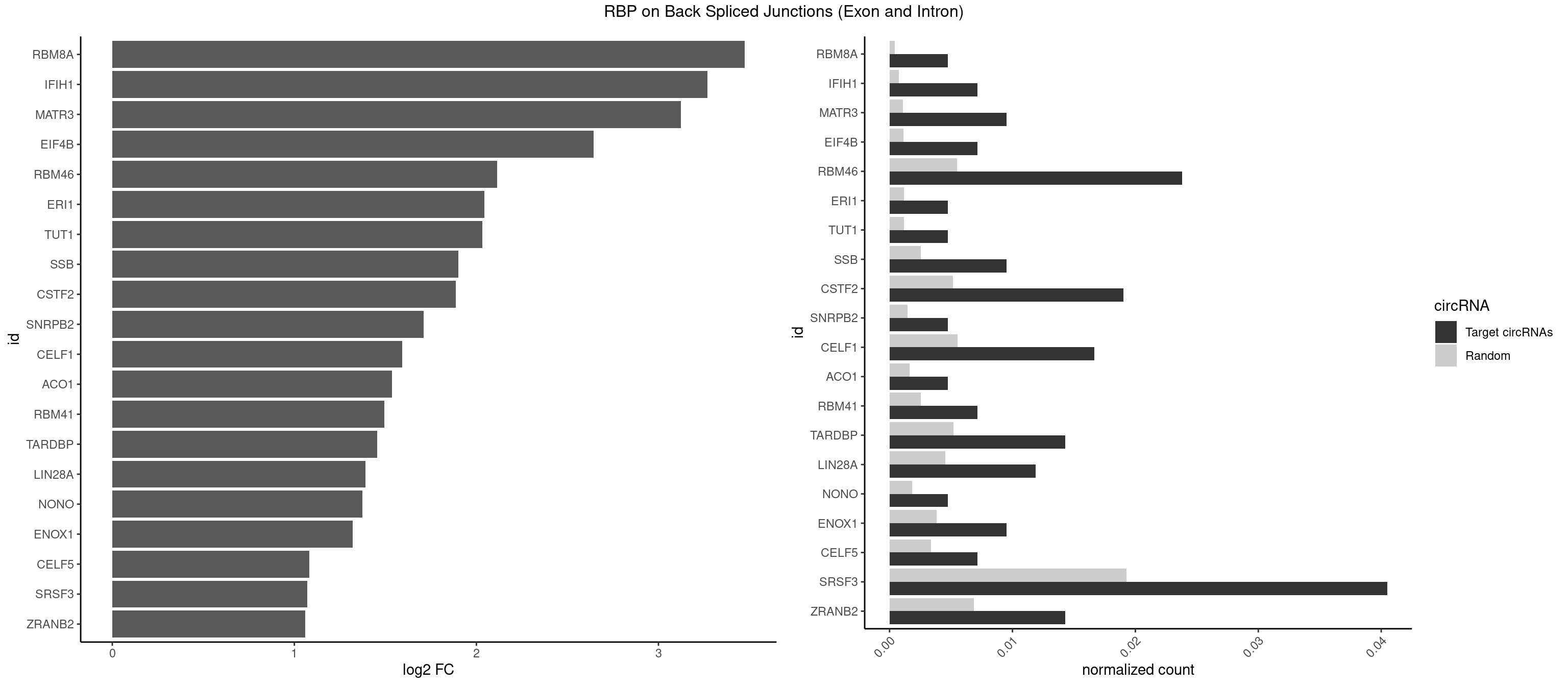
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM8A | 1 | 84 | 0.004761905 | 0.0004282547 | 3.474998 | UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| IFIH1 | 2 | 146 | 0.007142857 | 0.0007406288 | 3.269679 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| MATR3 | 3 | 216 | 0.009523810 | 0.0010933091 | 3.122837 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM46 | 9 | 1091 | 0.023809524 | 0.0055018138 | 2.113560 | AAUGAU,AUGAAU,AUGAUG,AUGAUU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| CSTF2 | 7 | 1022 | 0.019047619 | 0.0051541717 | 1.885798 | GUGUUU,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | AUUGCA | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| CELF1 | 6 | 1097 | 0.016666667 | 0.0055320435 | 1.591081 | GUUUGU,UGUGUU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACAU,UACAUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| TARDBP | 5 | 1034 | 0.014285714 | 0.0052146312 | 1.453936 | GAAUGA,GAAUGG,GUGAAU,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | GGAGUA,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AAUACA,CAUACA,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SRSF3 | 16 | 3825 | 0.040476190 | 0.0192765014 | 1.070230 | AACGAC,ACGACU,AUCUUC,AUUCAU,CUUCAG,CUUCAU,UACAAA,UCGACA,UCUUCA,UUCGAC,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| ZRANB2 | 5 | 1360 | 0.014285714 | 0.0068571141 | 1.058900 | AGGUUU,CGGUCU,GGUGGU,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.