circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000169402:+:7:6759083:6768766
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000169402:+:7:6759083:6768766 | ENSG00000169402 | ENST00000403107 | + | 7 | 6759084 | 6768766 | 703 | CUAUGAAGGGGAAAAGGUUCGUGGGCUGUAUGAGGGAGAAGGCUUCGCAGCCUUUCAAGGCGGUUGUACCUAUCGUGGUAUGUUUUCAGAAGGACUCAUGCAUGGACAGGGGACUUAUAUUUGGGCCGAUGGAUUAAAAUAUGAGGGCGACUUUGUGAAGAAUGUCCCGAUGAACCACGGCGUGUACACGUGGCCGGACGGCAGCAUGUAUGAAGGCGAAGUGGUCAACGGCAUGAGGAACGGAUUCGGGAUGUUCAAGUGCAGCACCCAGCCUGUGUCCUACAUCGGCCACUGGUGCAAUGGCAAGCGGCACGGGAAGGGCUCCAUUUAUUACAAUCAAGAGGGUACGUGUUGGUACGAGGGAGACUGGGUACAAAACAUCAAAAAGGGCUGGGGAAUAAGAUGUUAUAAAUCUGGAAAUAUAUACGAAGGCCAGUGGGAAGACAACAUGCGCCACGGGGAGGGGAGGAUGAGGUGGCUGACCACCAACGAAGAGUACACCGGGCGGUGGGAGAGGGGCAUCCAGAAUGGCUUUGGAACACACACAUGGUUUCUAAAGAGAAUCCGCAGUUCCCAGUAUCCUUUGAGAAAUGAAUACAUAGGGGAGUUUGUAAAUGGAUAUCGUCACGGACGUGGCAAGUUUUAUUAUGCCAGUGGAGCCAUGUAUGAUGGAGAAUGGGUUUCCAAUAAGAAACAUGGCAUGCUAUGAAGGGGAAAAGGUUCGUGGGCUGUAUGAGGGAGAAGGCUUCGCAG | circ |
| ENSG00000169402:+:7:6759083:6768766 | ENSG00000169402 | ENST00000403107 | + | 7 | 6759084 | 6768766 | 22 | AACAUGGCAUGCUAUGAAGGGG | bsj |
| ENSG00000169402:+:7:6759083:6768766 | ENSG00000169402 | ENST00000403107 | + | 7 | 6758884 | 6759093 | 210 | GAAAUGCCUUCAUUCCUCACCCUACUUUCAAACCCCUGGUAACCUCUUCUACUUUCUAGCUCUGUGUAUAUAUAUAUAUAUAUAUAUUUUUUUUUUUUCUGACUUCAUUCUUCCCAUUACAUAGUUAUAUAGUUUAACCCACUGUCACUUGGGUUUAUUCACAUUGUUCAUUACCUUGAAUUACCUUUAUUUCUUUGUAGCUAUGAAGGG | ie_up |
| ENSG00000169402:+:7:6759083:6768766 | ENSG00000169402 | ENST00000403107 | + | 7 | 6768757 | 6768966 | 210 | ACAUGGCAUGGUGAGUAUAGACCUGGGAGGAUCACAUUUCAAACAAUUAUGUAAAGCCAUUGAAAUUAUGUAAAGCAAUUGAAAUGUCCAAGGAUAAUUGCAGAUUUGUCUAUUCUGCUUUCAGAUCUAUCAGUUUUGAUUCAAAUAUUUGAAGCUUUGUUAGGUGCAUAUACAUUGAGGAUUAUUCUGUAUUGGUGCAUUGGCUCUUUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
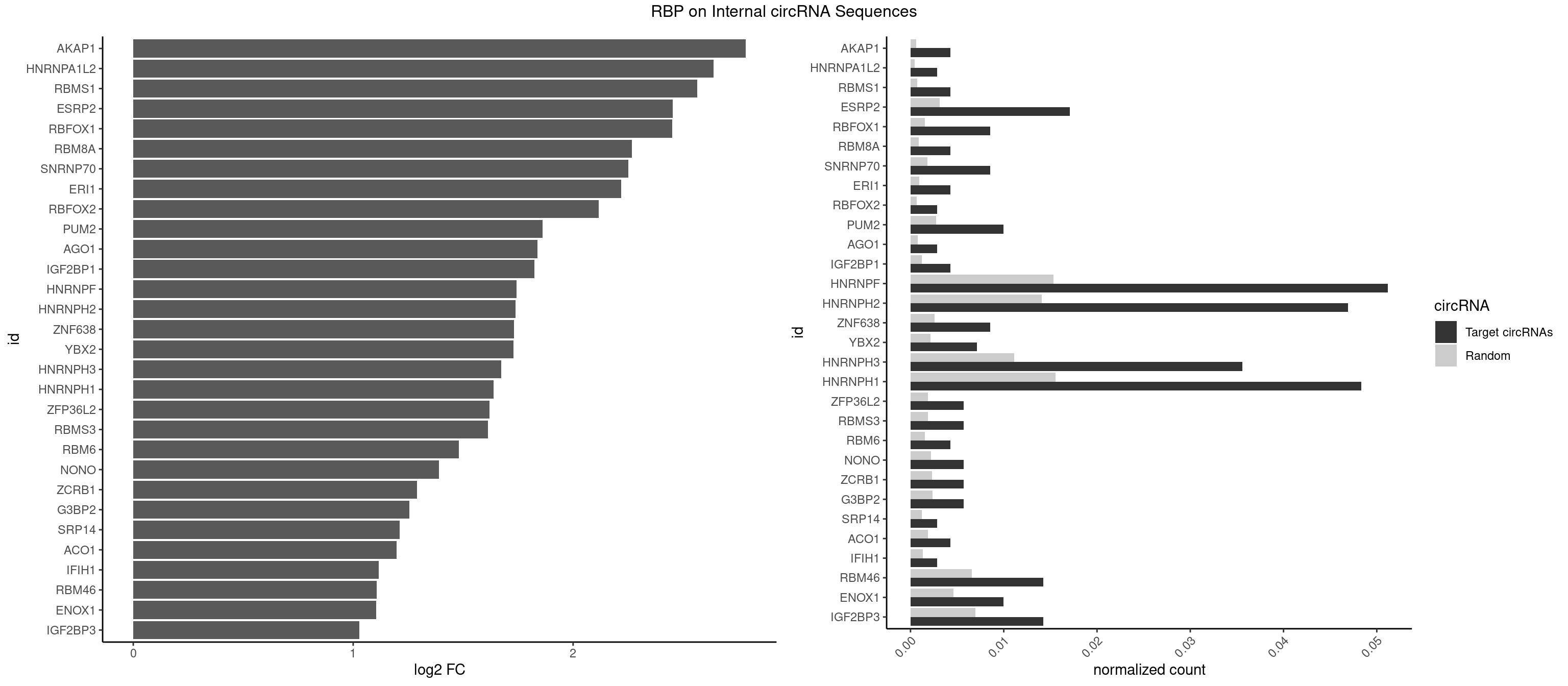
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| AKAP1 | 2 | 426 | 0.004267425 | 0.0006188101 | 2.785797 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| HNRNPA1L2 | 1 | 314 | 0.002844950 | 0.0004564992 | 2.639719 | AUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBMS1 | 2 | 497 | 0.004267425 | 0.0007217036 | 2.563888 | AUAUAC,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| ESRP2 | 11 | 2150 | 0.017069701 | 0.0031172377 | 2.453098 | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,UGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBFOX1 | 5 | 1077 | 0.008534851 | 0.0015622419 | 2.449748 | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBM8A | 2 | 611 | 0.004267425 | 0.0008869128 | 2.266502 | AUGCGC,UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| SNRNP70 | 5 | 1237 | 0.008534851 | 0.0017941145 | 2.250094 | AAUCAA,AUCAAG,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ERI1 | 2 | 632 | 0.004267425 | 0.0009173461 | 2.217828 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBFOX2 | 1 | 452 | 0.002844950 | 0.0006564894 | 2.115560 | UGCAUG | UGACUG,UGCAUG |
| PUM2 | 6 | 1890 | 0.009957326 | 0.0027404447 | 1.861348 | GUAAAU,UACAUA,UACAUC,UAUAUA,UGUAAA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| AGO1 | 1 | 548 | 0.002844950 | 0.0007956130 | 1.838265 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| IGF2BP1 | 2 | 831 | 0.004267425 | 0.0012057377 | 1.823450 | AGCACC,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPF | 35 | 10561 | 0.051209104 | 0.0153064921 | 1.742257 | AAGGCG,AAGGGG,AGGCGA,AGGGGA,CGAUGG,CGGGAU,CUGGGG,GAAGGG,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,UGGGAA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH2 | 32 | 9711 | 0.046941679 | 0.0140746688 | 1.737768 | AAGGCG,AAGGGG,AGGCGA,AGGGGA,CGAGGG,CGGGCG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCUG,GGGGAA,GGGGAG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| ZNF638 | 5 | 1773 | 0.008534851 | 0.0025708878 | 1.731099 | GGUUCG,GGUUGU,GUUCGU,GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| YBX2 | 4 | 1480 | 0.007112376 | 0.0021462711 | 1.728499 | AACAUC,ACAACA,ACAUCA,ACAUCG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| HNRNPH3 | 24 | 7688 | 0.035561878 | 0.0111429292 | 1.674203 | AGGGGA,CGAGGG,CGGGCG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCUG,GGGGAG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 33 | 10717 | 0.048364154 | 0.0155325680 | 1.638642 | AAGGCG,AAGGGG,AGGCGA,AGGGGA,CGAGGG,CGGGCG,CUGGGG,GAAGGG,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCUG,GGGGAA,GGGGAG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| ZFP36L2 | 3 | 1277 | 0.005689900 | 0.0018520827 | 1.619255 | AUUUAU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBMS3 | 3 | 1283 | 0.005689900 | 0.0018607779 | 1.612498 | AAUAUA,AUAUAU,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBM6 | 2 | 1054 | 0.004267425 | 0.0015289102 | 1.480862 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| NONO | 3 | 1498 | 0.005689900 | 0.0021723567 | 1.389142 | AGGAAC,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ZCRB1 | 3 | 1605 | 0.005689900 | 0.0023274215 | 1.289671 | GACUUA,GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| G3BP2 | 3 | 1644 | 0.005689900 | 0.0023839405 | 1.255055 | AGGAUG,GGAUGA,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SRP14 | 1 | 847 | 0.002844950 | 0.0012289250 | 1.211007 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ACO1 | 2 | 1283 | 0.004267425 | 0.0018607779 | 1.197460 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| IFIH1 | 1 | 904 | 0.002844950 | 0.0013115296 | 1.117153 | GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM46 | 9 | 4554 | 0.014224751 | 0.0066011240 | 1.107620 | AAUCAA,AAUGAA,AUCAAA,AUCAAG,AUGAAG,AUGAAU,AUGAUG,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ENOX1 | 6 | 3195 | 0.009957326 | 0.0046316558 | 1.104230 | AAGACA,AAUACA,AGUACA,GGACAG,UGGACA,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| IGF2BP3 | 9 | 4815 | 0.014224751 | 0.0069793662 | 1.027235 | AAAACA,AACACA,AAUACA,ACAAUC,ACACAC,CAAUCA,CACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
RBP on BSJ (Exon Only)
Plot
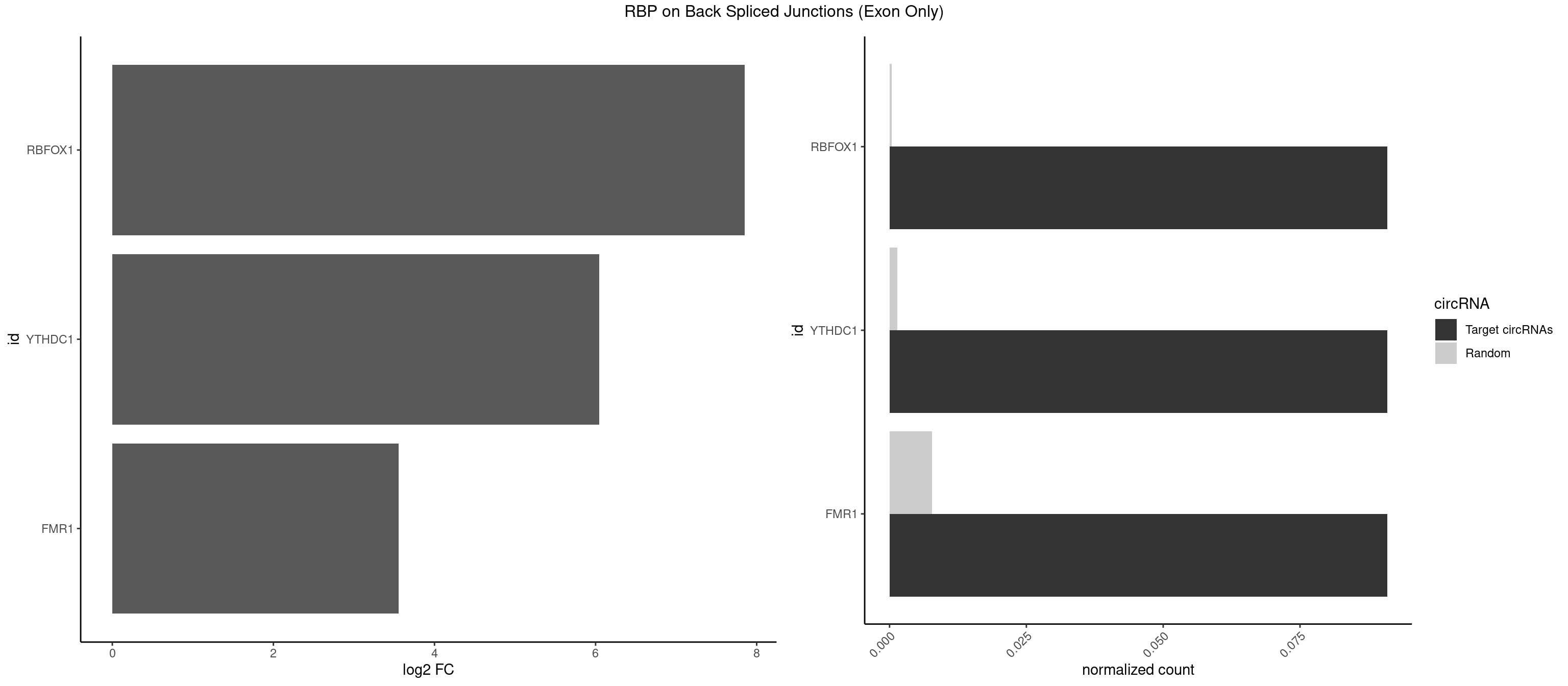
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX1 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GCAUGC | AGCAUG,GCAUGU,UGACUG,UGCAUG |
| YTHDC1 | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | GCAUGC | GAAUAC,GAGUAC,GCCUGC,GGCUGC,GGGUAC,UAAUAC,UAAUGC,UCAUAC,UCCUGC,UCGUGC,UGAUAC,UGCUGC,UGGUGC |
| FMR1 | 1 | 117 | 0.09090909 | 0.0077285827 | 3.556149 | GCUAUG | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
RBP on BSJ (Exon and Intron)
Plot
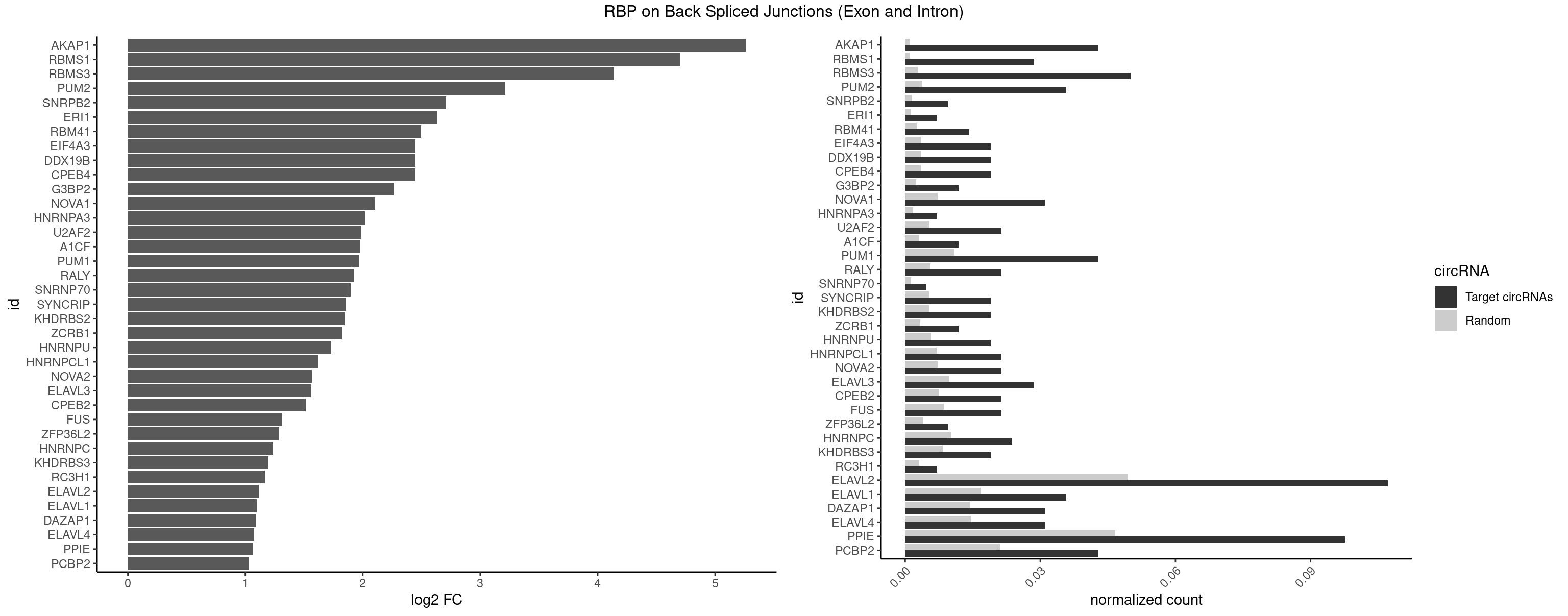
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| AKAP1 | 17 | 221 | 0.042857143 | 0.001118501 | 5.259898 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| RBMS1 | 11 | 217 | 0.028571429 | 0.001098347 | 4.701167 | AUAUAC,AUAUAG,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBMS3 | 20 | 562 | 0.050000000 | 0.002836558 | 4.139715 | AUAUAG,AUAUAU,CAUAUA,UAUAGA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| PUM2 | 14 | 764 | 0.035714286 | 0.003854293 | 3.211963 | GUAUAU,UACAUA,UAUAUA,UGUAAA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SNRPB2 | 3 | 288 | 0.009523810 | 0.001456066 | 2.709463 | AUUGCA,GUAUUG,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ERI1 | 2 | 228 | 0.007142857 | 0.001153769 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBM41 | 5 | 502 | 0.014285714 | 0.002534260 | 2.494937 | AUACAU,UACAUU,UACUUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| CPEB4 | 7 | 691 | 0.019047619 | 0.003486497 | 2.449760 | UUUUUU | UUUUUU |
| DDX19B | 7 | 691 | 0.019047619 | 0.003486497 | 2.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 7 | 691 | 0.019047619 | 0.003486497 | 2.449760 | UUUUUU | UUUUUU |
| G3BP2 | 4 | 490 | 0.011904762 | 0.002473801 | 2.266737 | AGGAUA,AGGAUU,GGAUAA,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| NOVA1 | 12 | 1428 | 0.030952381 | 0.007199718 | 2.104038 | UCAUUC,UUCAUU,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.001763402 | 2.018140 | CAAGGA,CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| U2AF2 | 8 | 1071 | 0.021428571 | 0.005401048 | 1.988224 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| A1CF | 4 | 598 | 0.011904762 | 0.003017936 | 1.979904 | AGUAUA,AUAAUU,AUCAGU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PUM1 | 17 | 2172 | 0.042857143 | 0.010948206 | 1.968841 | AAUAUU,GUAUAU,UACAUA,UAUAUA,UGUAAA,UGUAUA,UGUCCA,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| RALY | 8 | 1117 | 0.021428571 | 0.005632809 | 1.927609 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.001279726 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| SYNCRIP | 7 | 1041 | 0.019047619 | 0.005249899 | 1.859249 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 7 | 1051 | 0.019047619 | 0.005300282 | 1.845470 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.003360540 | 1.824774 | GAAUUA,GGAUUA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPU | 7 | 1136 | 0.019047619 | 0.005728537 | 1.733372 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPCL1 | 8 | 1381 | 0.021428571 | 0.006962918 | 1.621772 | AUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| NOVA2 | 8 | 1434 | 0.021428571 | 0.007229948 | 1.567479 | GAUCAC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| ELAVL3 | 11 | 1928 | 0.028571429 | 0.009718863 | 1.555714 | UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| CPEB2 | 8 | 1487 | 0.021428571 | 0.007496977 | 1.515155 | AUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| FUS | 8 | 1711 | 0.021428571 | 0.008625554 | 1.312847 | UGGUGA,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.003904676 | 1.286336 | UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPC | 9 | 2006 | 0.023809524 | 0.010111850 | 1.235492 | AUUUUU,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS3 | 7 | 1646 | 0.019047619 | 0.008298065 | 1.198764 | UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.003184200 | 1.165570 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ELAVL2 | 44 | 9826 | 0.107142857 | 0.049511286 | 1.113706 | AUAUUU,AUUAUU,AUUUUU,CUUUCU,GUAUUG,UACUUU,UAGUUA,UAUAUA,UAUAUU,UAUUUG,UAUUUU,UCUUUU,UGUAUU,UUAUAU,UUAUGU,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUUAUU,UUUCUU,UUUGAU,UUUUUC,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL1 | 14 | 3309 | 0.035714286 | 0.016676743 | 1.098664 | UAGUUU,UAUUUU,UUAUUU,UUGAUU,UUUAUU,UUUGUU,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| DAZAP1 | 12 | 2878 | 0.030952381 | 0.014505240 | 1.093476 | AGGAUA,AGUAUA,AGUUUA,UAGUUA,UAGUUU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ELAVL4 | 12 | 2916 | 0.030952381 | 0.014696695 | 1.074559 | UAUUUU,UUAUUU,UUUAUU,UUUGUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| PPIE | 40 | 9262 | 0.097619048 | 0.046669690 | 1.064677 | AAAUAU,AAAUUA,AAUAUU,AAUUAU,AUAAUU,AUAUAU,AUAUUU,AUUAUU,AUUUUU,UAUAUA,UAUAUU,UAUUUU,UUAUAU,UUAUUU,UUUAUU,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| PCBP2 | 17 | 4164 | 0.042857143 | 0.020984482 | 1.030213 | AAAUUA,AAUUAC,ACCCUA,AUUACC,AUUCAC,CAAUUA,CCAUUA,CUUCCC,UAACCC,UUUUUU | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.