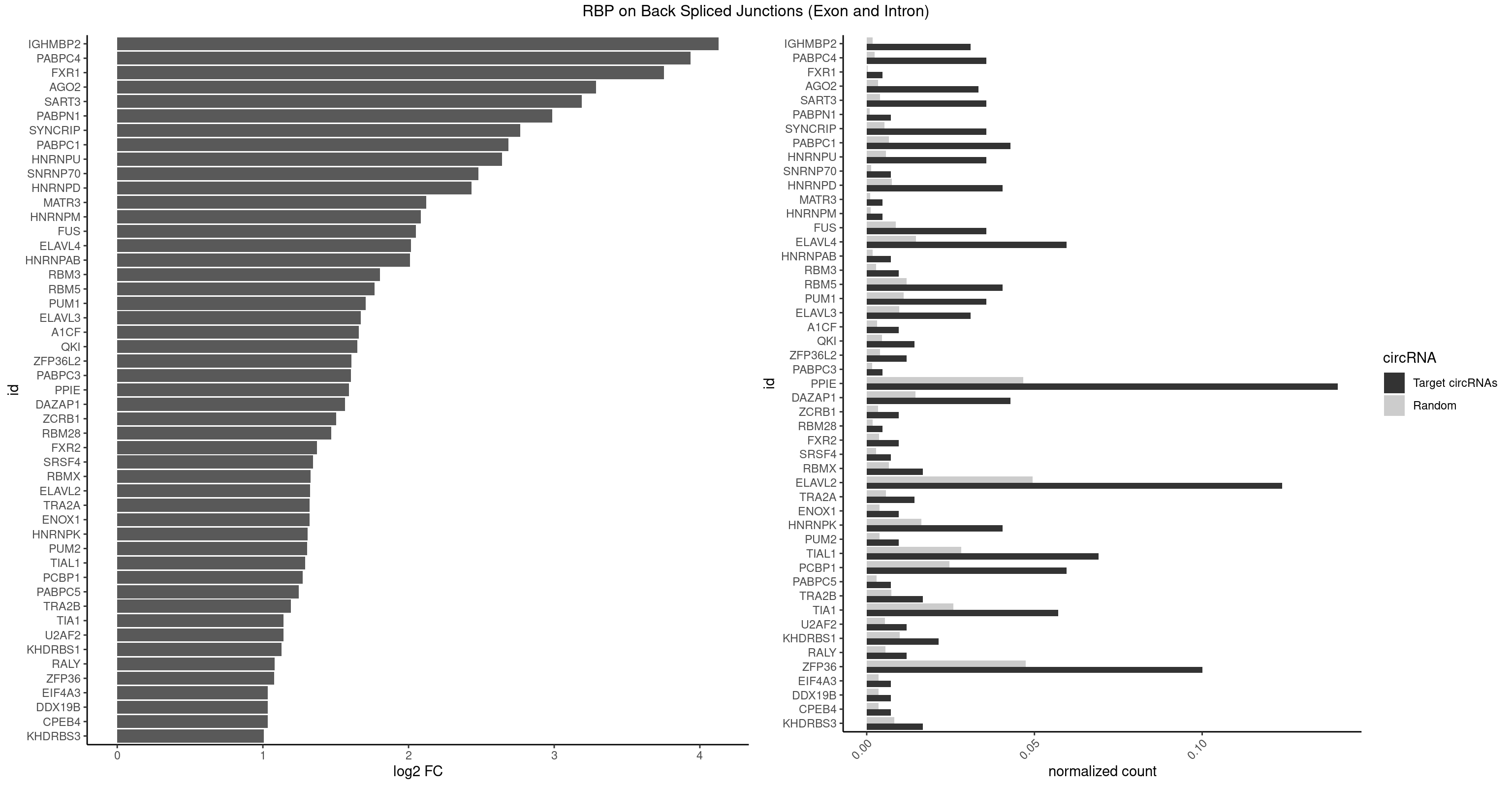
| IGHMBP2 |
12 |
350 |
0.030952381 |
0.0017684401 |
4.129501 |
AAAAAA |
AAAAAA |
| PABPC4 |
14 |
462 |
0.035714286 |
0.0023327287 |
3.936411 |
AAAAAA,AAAAAG |
AAAAAA,AAAAAG |
| FXR1 |
1 |
69 |
0.004761905 |
0.0003526804 |
3.755106 |
AUGACA |
ACGACA,ACGACG,AUGACA,AUGACG |
| AGO2 |
13 |
677 |
0.033333333 |
0.0034159613 |
3.286602 |
AAAAAA,UAAAGU |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SART3 |
14 |
777 |
0.035714286 |
0.0039197904 |
3.187653 |
AAAAAA,AGAAAA,GAAAAC |
AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPN1 |
2 |
178 |
0.007142857 |
0.0009018541 |
2.985535 |
AAAAGA,AGAAGA |
AAAAGA,AGAAGA |
| SYNCRIP |
14 |
1041 |
0.035714286 |
0.0052498992 |
2.766140 |
AAAAAA,UUUUUU |
AAAAAA,UUUUUU |
| PABPC1 |
17 |
1321 |
0.042857143 |
0.0066606207 |
2.685807 |
AAAAAA,ACAAAU,AGAAAA,CAAAUA,CAAAUC,GAAAAC |
AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| HNRNPU |
14 |
1136 |
0.035714286 |
0.0057285369 |
2.640263 |
AAAAAA,UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| SNRNP70 |
2 |
253 |
0.007142857 |
0.0012797259 |
2.480666 |
GUUCAA,UUCAAG |
AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPD |
16 |
1488 |
0.040476190 |
0.0075020153 |
2.431723 |
AAAAAA,UAUUUA,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| MATR3 |
1 |
216 |
0.004761905 |
0.0010933091 |
2.122837 |
AAUCUU |
AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| HNRNPM |
1 |
222 |
0.004761905 |
0.0011235389 |
2.083489 |
AAGGAA |
AAGGAA,GAAGGA,GGGGGG |
| FUS |
14 |
1711 |
0.035714286 |
0.0086255542 |
2.049812 |
AAAAAA,UUUUUU |
AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ELAVL4 |
24 |
2916 |
0.059523810 |
0.0146966949 |
2.017975 |
AAAAAA,UAUUUA,UAUUUU,UCUAAU,UUAUUU,UUUAUU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPAB |
2 |
351 |
0.007142857 |
0.0017734784 |
2.009919 |
AAGACA,GACAAA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 |
3 |
541 |
0.009523810 |
0.0027307537 |
1.802240 |
AAAACU,GAAACU |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBM5 |
16 |
2359 |
0.040476190 |
0.0118903668 |
1.767280 |
AAAAAA,AAGGAA,UCUUCU,UUCUCU |
AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| PUM1 |
14 |
2172 |
0.035714286 |
0.0109482064 |
1.705807 |
AAUAUU,AGAAUU,GAAUUG,GUAAAU,GUAAUA,UAAAUA,UAAUAU,UGUAAA,UUAAUG,UUUAAU |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ELAVL3 |
12 |
1928 |
0.030952381 |
0.0097188634 |
1.671191 |
AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| A1CF |
3 |
598 |
0.009523810 |
0.0030179363 |
1.657976 |
UAAUUA,UUAAUU |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| QKI |
5 |
904 |
0.014285714 |
0.0045596534 |
1.647577 |
ACUCAU,CUACUC,UACUCA,UCAUAU,UCUAAU |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ZFP36L2 |
4 |
774 |
0.011904762 |
0.0039046755 |
1.608264 |
UAUUUA,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PABPC3 |
1 |
310 |
0.004761905 |
0.0015669085 |
1.603618 |
GAAAAC |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| PPIE |
58 |
9262 |
0.140476190 |
0.0466696896 |
1.589768 |
AAAAAA,AAAUAA,AAAUAU,AAUAUU,AAUUAA,AAUUUU,AUAAUA,AUAUUU,AUUAAA,AUUAUA,AUUUAA,AUUUUA,AUUUUU,UAAAAA,UAAAUA,UAAUAU,UAAUUA,UAAUUU,UAUAAU,UAUUUA,UAUUUU,UUAAAA,UUAAUA,UUAAUU,UUAUAA,UUAUUU,UUUAAU,UUUAUU,UUUUAA,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| DAZAP1 |
17 |
2878 |
0.042857143 |
0.0145052398 |
1.562962 |
AAAAAA,AGGAAG,AGUUUA,UAGUUA,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ZCRB1 |
3 |
666 |
0.009523810 |
0.0033605401 |
1.502846 |
AAUUAA,AUUUAA,GAUUAA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM28 |
1 |
340 |
0.004761905 |
0.0017180572 |
1.470761 |
GUGUAG |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| FXR2 |
3 |
730 |
0.009523810 |
0.0036829907 |
1.370661 |
AGACAG,GACAAA,UGACAA |
AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| SRSF4 |
2 |
558 |
0.007142857 |
0.0028164047 |
1.342647 |
AGAAGA,AGGAAG |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBMX |
6 |
1316 |
0.016666667 |
0.0066354293 |
1.328704 |
AAGGAA,AAGUAA,AGGAAG,AGUAAC,AUCCCA,UAAGAC |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ELAVL2 |
51 |
9826 |
0.123809524 |
0.0495112858 |
1.322293 |
AAUUUU,AUAUUU,AUUUAA,AUUUUA,AUUUUC,AUUUUU,CUUUCU,CUUUUA,GUUUUA,GUUUUC,GUUUUU,UAAUUU,UAGUUA,UAUUGU,UAUUUA,UAUUUU,UCUUUU,UGUAUU,UUAAUU,UUACUU,UUAGUU,UUAUGU,UUAUUG,UUAUUU,UUCUUU,UUUAAU,UUUACU,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUUAA,UUUUUA,UUUUUC,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TRA2A |
5 |
1133 |
0.014285714 |
0.0057134220 |
1.322146 |
AAAGAA,AAGAAA,AGAAGA,AGGAAG,GAAGAG |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ENOX1 |
3 |
756 |
0.009523810 |
0.0038139863 |
1.320239 |
AAGACA,AAUACA,AGACAG |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPK |
16 |
3244 |
0.040476190 |
0.0163492543 |
1.307849 |
AAAAAA,UCCCAA,UCCCAC,UUUUUU |
AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| PUM2 |
3 |
764 |
0.009523810 |
0.0038542926 |
1.305073 |
GUAAAU,UAAAUA,UGUAAA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| TIAL1 |
28 |
5597 |
0.069047619 |
0.0282043531 |
1.291674 |
AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUU,CAGUUU,CUUUUA,GUUUUC,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAA,UUUUCA,UUUUUA,UUUUUC,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PCBP1 |
24 |
4891 |
0.059523810 |
0.0246473196 |
1.272036 |
AAAAAA,AAACCA,AAAUUC,AACCAA,AAUUAA,AUUAAA,CAUUCC,CCCUUA,CCUUUC,UUUUUU |
AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| PABPC5 |
2 |
596 |
0.007142857 |
0.0030078597 |
1.247764 |
AGAAAA,GAAAUU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TRA2B |
6 |
1447 |
0.016666667 |
0.0072954454 |
1.191898 |
AAAGAA,AAGGAA,AAUAAG,AGAAGA,AGGAAG,AUAAGA |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| TIA1 |
23 |
5140 |
0.057142857 |
0.0259018541 |
1.141518 |
AUUUUC,AUUUUU,CCUUUC,CUUUUA,GUUUUA,GUUUUC,GUUUUU,UAUUUU,UCCUUU,UUAUUU,UUUAUU,UUUUCU,UUUUUA,UUUUUC,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| U2AF2 |
4 |
1071 |
0.011904762 |
0.0054010480 |
1.140228 |
UUUUUC,UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| KHDRBS1 |
8 |
1945 |
0.021428571 |
0.0098045143 |
1.128018 |
AUUUAA,GAAAAC,UAAAAA,UUAAAA,UUUUAC,UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RALY |
4 |
1117 |
0.011904762 |
0.0056328094 |
1.079612 |
UUUUUC,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| ZFP36 |
41 |
9408 |
0.100000000 |
0.0474052801 |
1.076880 |
AAAAAA,AAAAAG,AAAAGA,AAAAGG,AAAGAA,AAAGGA,AAAUAA,AAGAAA,AAGGAA,AAGUAA,AAUAAG,ACAAAU,ACUUCC,AUUUAA,CAAAUA,CAAGUA,CCUUUC,UAAAUA,UAUUUA,UCCUUU,UCUUCU,UCUUUU,UUAUUU,UUUAAU,UUUAUU |
AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
| CPEB4 |
2 |
691 |
0.007142857 |
0.0034864974 |
1.034723 |
UUUUUU |
UUUUUU |
| DDX19B |
2 |
691 |
0.007142857 |
0.0034864974 |
1.034723 |
UUUUUU |
UUUUUU |
| EIF4A3 |
2 |
691 |
0.007142857 |
0.0034864974 |
1.034723 |
UUUUUU |
UUUUUU |
| KHDRBS3 |
6 |
1646 |
0.016666667 |
0.0082980653 |
1.006119 |
AAAUAA,AUUAAA,UAAAUA,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |