circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000131018:-:6:152201823:152215060
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000131018:-:6:152201823:152215060 | ENSG00000131018 | ENST00000367255 | - | 6 | 152201824 | 152215060 | 954 | GGACAGAUGUUAAAAUUCAGCAGCAUGGCUCCAGAUUUAGACCGUCUAAAUGAGCUUGGAUAUAGGUUACCCUUGAAUGAUAAGGAAAUCAAAAGAAUGCAGAAUCUGAACCGCCAUUGGUCUCUGAUCUCCUCUCAGACUACAGAAAGAUUCAGCAAGUUGCAGUCAUUUUUGCUACAACAUCAGACUUUCUUGGAAAAAUGUGAAACAUGGAUGGAAUUCCUAGUUCAGACAGAACAAAAGUUAGCAGUAGAGAUUUCAGGAAAUUAUCAGCACCUUUUGGAACAGCAGAGAGCACACGAGUUGUUUCAAGCCGAGAUGUUCAGUCGUCAGCAGAUUUUGCACUCAAUCAUUAUUGAUGGGCAACGUCUUCUAGAACAAGGUCAAGUUGAUGACAGGGAUGAAUUCAACCUGAAAUUGACACUCCUCAGUAAUCAAUGGCAGGGAGUGAUUCGCAGGGCCCAGCAGAGGCGGGGGAUCAUUGACAGCCAGAUUCGCCAGUGGCAGCGCUAUAGGGAGAUGGCAGAAAAGCUUCGUAAAUGGUUGGUUGAAGUGUCCUACCUCCCCAUGAGUGGUCUCGGAAGUGUUCCUAUACCACUGCAACAAGCAAGGACCCUCUUUGAUGAAGUGCAGUUCAAAGAAAAAGUGUUUCUGCGGCAACAAGGCAGCUACAUCCUGACUGUGGAGGCUGGCAAGCAACUCCUUCUCUCGGCGGACAGUGGCGCUGAGGCCGCCUUGCAGGCCGAACUCGCUGAAAUCCAAGAGAAAUGGAAAUCAGCCAGCAUGCGGCUGGAAGAACAGAAGAAAAAACUAGCCUUCUUGUUGAAAGACUGGGAAAAAUGUGAGAAAGGAAUAGCAGAUUCCCUGGAGAAACUACGAACUUUCAAAAAGAAGCUUUCGCAGUCUCUCCCGGAUCACCAUGAAGAGCUCCAUGCAGAACAAAUGCGUUGCAAGGGACAGAUGUUAAAAUUCAGCAGCAUGGCUCCAGAUUUAGACCGUCUAAA | circ |
| ENSG00000131018:-:6:152201823:152215060 | ENSG00000131018 | ENST00000367255 | - | 6 | 152201824 | 152215060 | 22 | UGCGUUGCAAGGGACAGAUGUU | bsj |
| ENSG00000131018:-:6:152201823:152215060 | ENSG00000131018 | ENST00000367255 | - | 6 | 152215051 | 152215260 | 210 | GCAGCAGCAGUUUAGACUGAAAAGUAGAUUUGGCUUUUCGCUAUUCGUACCUAGAGAUUGUAAGUAAGUGGCCUAGAAAUGCAGAGGCCACUAGACCGAAGACCAGCUACUUCCUGAAAUCAGAAAUUCACUGCGCACUUUGAAAAAGUUUUGACUUUUUUGACCAUGGUGCUAAACCUACUUUUAAAUCUUCCUUCUAGGGACAGAUGU | ie_up |
| ENSG00000131018:-:6:152201823:152215060 | ENSG00000131018 | ENST00000367255 | - | 6 | 152201624 | 152201833 | 210 | GCGUUGCAAGGUAAAUUACUGAACUGAGAUUUUCACCGUCACCUGUGUGCCUCUGCUGGAAUGUCUUUGGGGCCAUAAUCCAGUCAAACUCUUAGGUUAUCUACAUGCAGAUACAAGAUCCUAGUAUAUGACAUAAGGACAGGCAACUCAGGUGAAACUUGUAUGGCUGAUUUAUUGCAGUUGAUGAAAUUAUAGACUAGUUUAUGGAUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
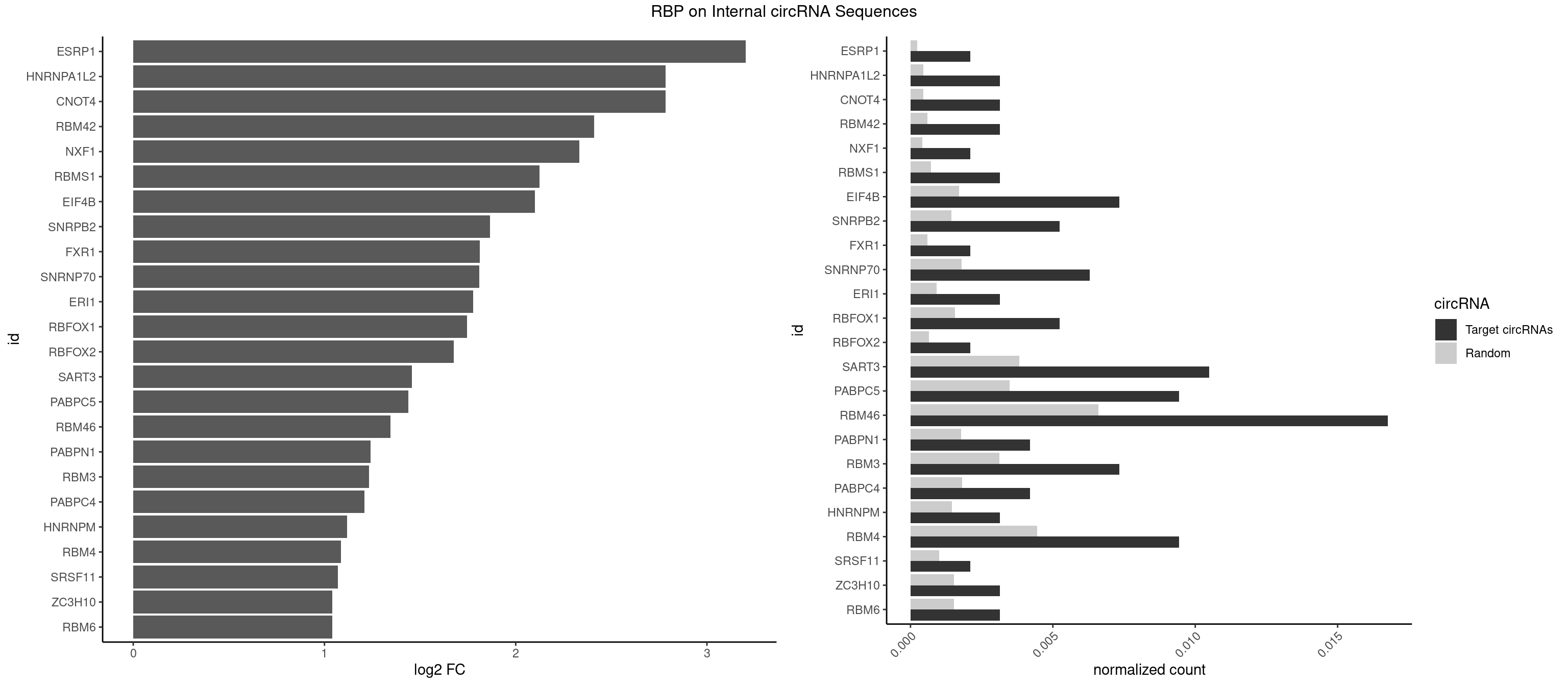
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.002096436 | 0.0002275250 | 3.203842 | AGGGAU | AGGGAU |
| CNOT4 | 2 | 314 | 0.003144654 | 0.0004564992 | 2.784217 | GACAGA | GACAGA |
| HNRNPA1L2 | 2 | 314 | 0.003144654 | 0.0004564992 | 2.784217 | AUAGGG,UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBM42 | 2 | 407 | 0.003144654 | 0.0005912752 | 2.411000 | AACUAC,ACUACG | AACUAA,AACUAC,ACUAAG,ACUACG |
| NXF1 | 1 | 286 | 0.002096436 | 0.0004159215 | 2.333556 | AACCUG | AACCUG |
| RBMS1 | 2 | 497 | 0.003144654 | 0.0007217036 | 2.123423 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| EIF4B | 6 | 1179 | 0.007337526 | 0.0017100607 | 2.101246 | CUCGGA,CUUGGA,UCGGAA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SNRPB2 | 4 | 991 | 0.005241090 | 0.0014376103 | 1.866194 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| FXR1 | 1 | 411 | 0.002096436 | 0.0005970720 | 1.811962 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SNRNP70 | 5 | 1237 | 0.006289308 | 0.0017941145 | 1.809629 | AAUCAA,AUUCAA,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ERI1 | 2 | 632 | 0.003144654 | 0.0009173461 | 1.777363 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBFOX1 | 4 | 1077 | 0.005241090 | 0.0015622419 | 1.746249 | AGCAUG,GCAUGC,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBFOX2 | 1 | 452 | 0.002096436 | 0.0006564894 | 1.675095 | UGACUG | UGACUG,UGCAUG |
| SART3 | 9 | 2634 | 0.010482180 | 0.0038186524 | 1.456803 | AAAAAA,AAAAAC,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPC5 | 8 | 2400 | 0.009433962 | 0.0034795387 | 1.438968 | AGAAAA,AGAAAG,AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBM46 | 15 | 4554 | 0.016771488 | 0.0066011240 | 1.345227 | AAUCAA,AAUCAU,AAUGAU,AUCAAA,AUCAAU,AUCAUU,AUGAAG,AUGAAU,AUGAUA,GAUCAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| PABPN1 | 3 | 1222 | 0.004192872 | 0.0017723764 | 1.242254 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBM3 | 6 | 2152 | 0.007337526 | 0.0031201361 | 1.233685 | AAAACU,AAACUA,AAGACU,AGACUA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PABPC4 | 3 | 1251 | 0.004192872 | 0.0018144033 | 1.208444 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| HNRNPM | 2 | 999 | 0.003144654 | 0.0014492040 | 1.117641 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| RBM4 | 8 | 3065 | 0.009433962 | 0.0044432593 | 1.086245 | CCUCUU,CCUUCU,CUCUUU,CUUCUU,UCCUUC,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| SRSF11 | 1 | 688 | 0.002096436 | 0.0009985015 | 1.070102 | AAGAAG | AAGAAG |
| ZC3H10 | 2 | 1053 | 0.003144654 | 0.0015274610 | 1.041766 | CAGCGC,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBM6 | 2 | 1054 | 0.003144654 | 0.0015289102 | 1.040398 | AAUCCA,AUCCAA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
RBP on BSJ (Exon Only)
Plot
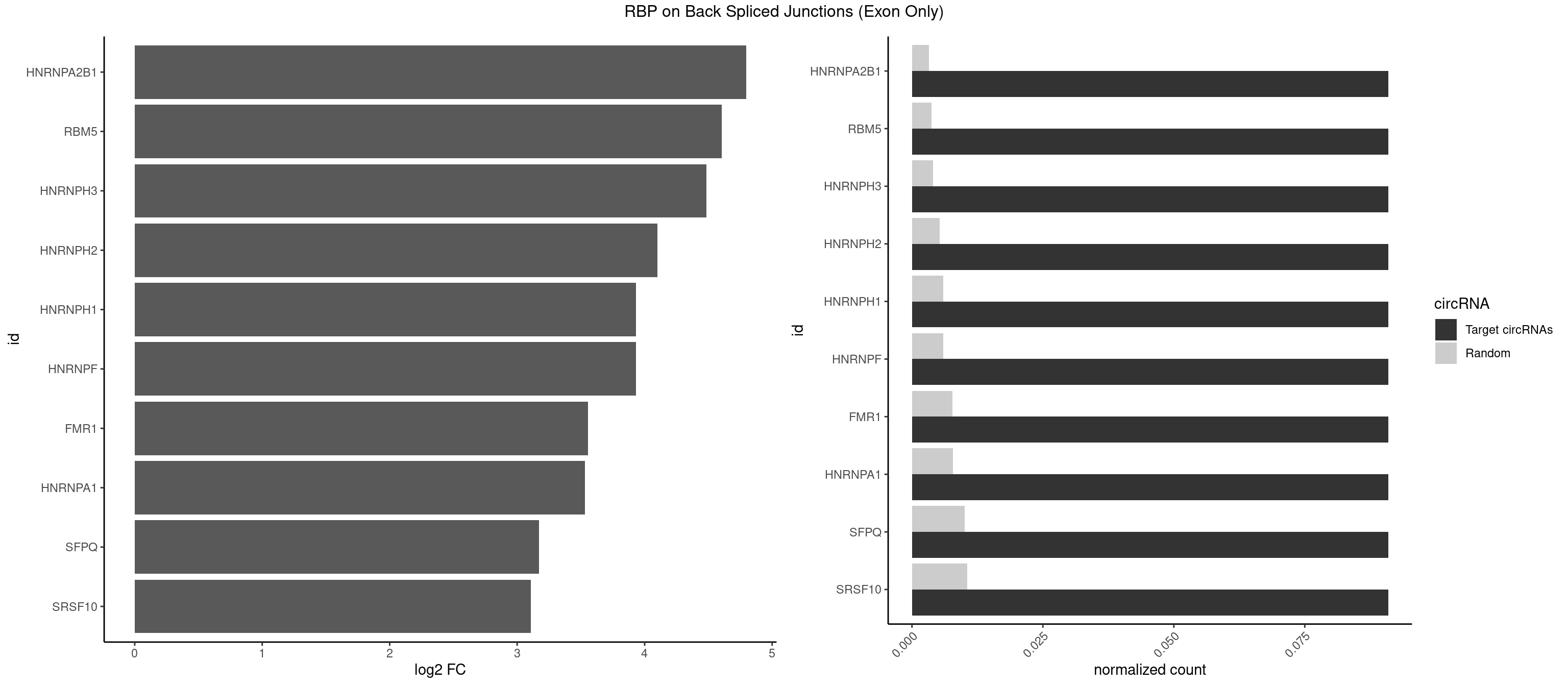
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.003274823 | 4.794936 | CAAGGG | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| RBM5 | 1 | 56 | 0.09090909 | 0.003733298 | 4.605902 | CAAGGG | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
| HNRNPH3 | 1 | 61 | 0.09090909 | 0.004060781 | 4.484596 | AAGGGA | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| HNRNPH2 | 1 | 80 | 0.09090909 | 0.005305214 | 4.098942 | AAGGGA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPF | 1 | 90 | 0.09090909 | 0.005960178 | 3.930997 | AAGGGA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 1 | 90 | 0.09090909 | 0.005960178 | 3.930997 | AAGGGA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| FMR1 | 1 | 117 | 0.09090909 | 0.007728583 | 3.556149 | AAGGGA | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.007859576 | 3.531901 | AGGGAC | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
| SFPQ | 1 | 153 | 0.09090909 | 0.010086455 | 3.172005 | AAGGGA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF10 | 1 | 160 | 0.09090909 | 0.010544931 | 3.107875 | AAGGGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon and Intron)
Plot
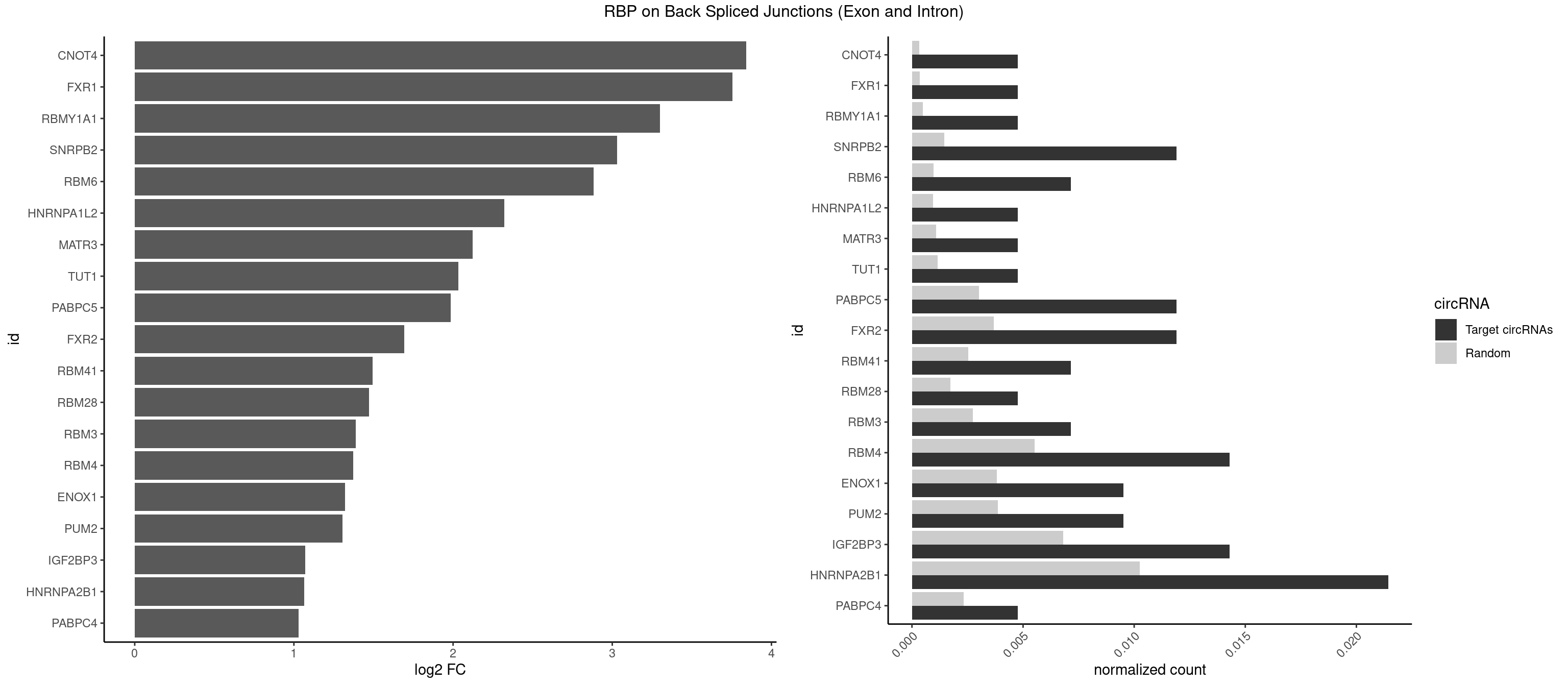
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | ACAAGA | ACAAGA,CAAGAC |
| SNRPB2 | 4 | 288 | 0.011904762 | 0.0014560661 | 3.031391 | AUUGCA,UAUUGC,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC5 | 4 | 596 | 0.011904762 | 0.0030078597 | 1.984730 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| FXR2 | 4 | 730 | 0.011904762 | 0.0036829907 | 1.692589 | GACAGA,GACAGG,GGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACAUG,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AGACUA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBM4 | 5 | 1094 | 0.014285714 | 0.0055169287 | 1.372636 | CCUUCU,CUUCCU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AGGACA,GGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | GUAAAU,GUAGAU,GUAUAU | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAACUC,AAAUCA,AAAUUC,AACUCA,AAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| HNRNPA2B1 | 8 | 2033 | 0.021428571 | 0.0102478839 | 1.064210 | ACUAGA,AGACUA,CUAGAC,GACUAG,GGGGCC,UAGACU,UAGGGA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.