circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000112699:-:6:1726415:1742586
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000112699:-:6:1726415:1742586 | ENSG00000112699 | ENST00000380805 | - | 6 | 1726416 | 1742586 | 216 | GCUAUGUGGUUGAUGUUGCAGAAUGAUGAGCCGGAGGACUUCGUUAUAGCUACUGGGGAGGUCCAUAGUGUCCGGGAAUUUGUCGAGAAAUCAUUCUUGCACAUUGGAAAAACCAUUGUGUGGGAAGGAAAGAAUGAAAAUGAAGUGGGCAGAUGUAAAGAGACCGGCAAAGUUCACGUGACUGUGGAUCUCAAGUACUACCGGCCAACUGAAGUGGCUAUGUGGUUGAUGUUGCAGAAUGAUGAGCCGGAGGACUUCGUUAUAGC | circ |
| ENSG00000112699:-:6:1726415:1742586 | ENSG00000112699 | ENST00000380805 | - | 6 | 1726416 | 1742586 | 22 | CAACUGAAGUGGCUAUGUGGUU | bsj |
| ENSG00000112699:-:6:1726415:1742586 | ENSG00000112699 | ENST00000380805 | - | 6 | 1742577 | 1742786 | 210 | GGCAUCCACACCAAGAAGUUCCUCAUGUUAUUCGGAUACUCAGUAAAAGUUGGGAGCCAUUGCUGUUGUGCUUAGAAUAUGCUAUGAAAUUCAUGCUCAUGUUCCAGCUGUCGAUGUCCAUAUCUGAUUAUGUGCACUGGAAAACAAUGUGUGGCCACUGAGUGUGACUUCAAAGUGAACUUGCCUCUCUUUCCCUCUAGGCUAUGUGGU | ie_up |
| ENSG00000112699:-:6:1726415:1742586 | ENSG00000112699 | ENST00000380805 | - | 6 | 1726216 | 1726425 | 210 | AACUGAAGUGGUAAGGACUCUCUGGCCACCCAGUGCGUGGCCACAUUUGGCUGGCUGGCUGGGCAAAUGCGCUCAUGGAAUGCCUGGUGCCAGCCUUUGGAGCUGUCAGUUCAGCUCCCUGGCAUCCUUCAGCGUGCAUUUCCAAAAUCAGAACACAGUGAAUUCAAUUUGAGAUUAUCAUUCUUACCACAAACUUCAUUGCUCUAUAAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
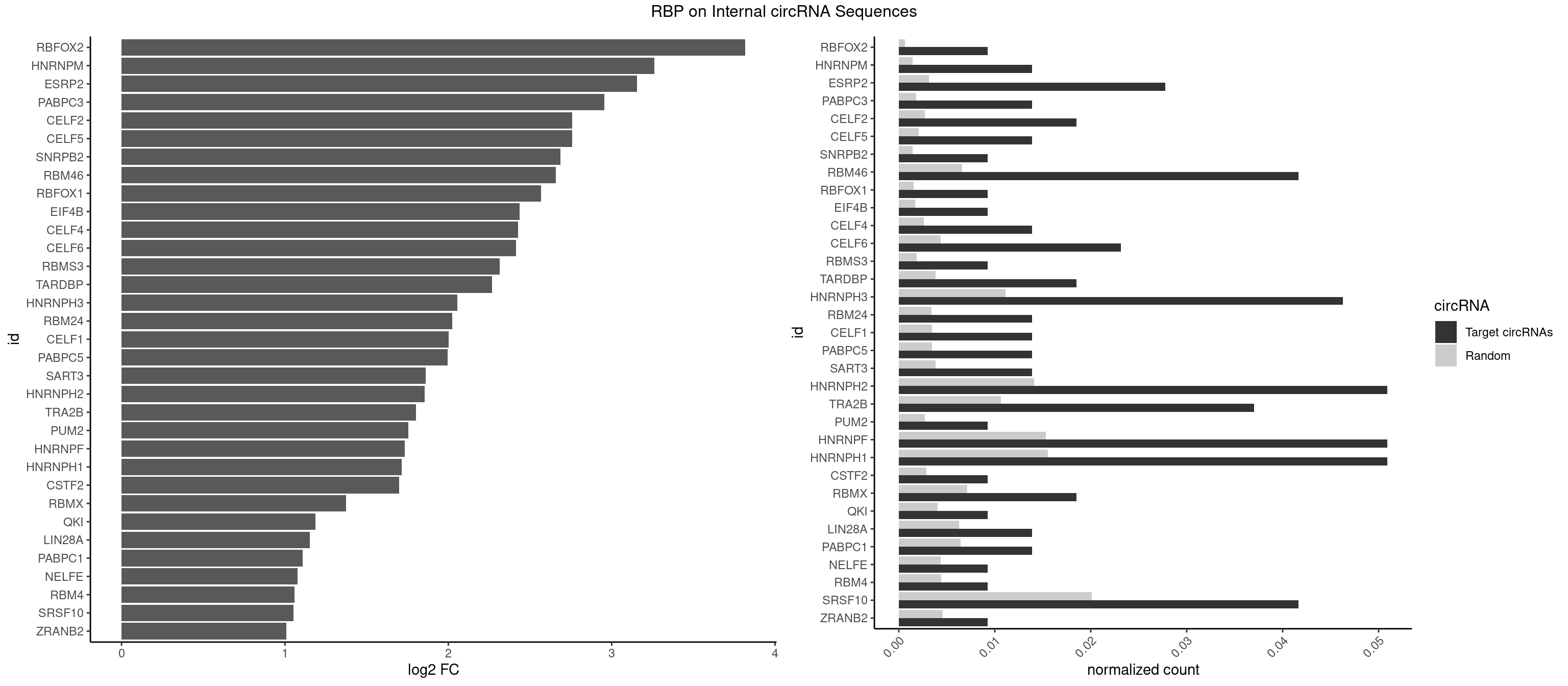
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 1 | 452 | 0.009259259 | 0.0006564894 | 3.818053 | UGACUG | UGACUG,UGCAUG |
| HNRNPM | 2 | 999 | 0.013888889 | 0.0014492040 | 3.260599 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ESRP2 | 5 | 2150 | 0.027777778 | 0.0031172377 | 3.155591 | GGGAAG,GGGAAU,GGGGAG,UGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| PABPC3 | 2 | 1234 | 0.013888889 | 0.0017897669 | 2.956088 | AAAAAC,AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| CELF2 | 3 | 1886 | 0.018518519 | 0.0027346479 | 2.759542 | UAUGUG,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 2 | 1415 | 0.013888889 | 0.0020520728 | 2.758777 | GUGUGG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SNRPB2 | 1 | 991 | 0.009259259 | 0.0014376103 | 2.687224 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM46 | 8 | 4554 | 0.041666667 | 0.0066011240 | 2.658110 | AAUCAU,AAUGAA,AAUGAU,AUCAUU,AUGAAA,AUGAAG,AUGAUG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBFOX1 | 1 | 1077 | 0.009259259 | 0.0015622419 | 2.567279 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| EIF4B | 1 | 1179 | 0.009259259 | 0.0017100607 | 2.436849 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| CELF4 | 2 | 1782 | 0.013888889 | 0.0025839306 | 2.426292 | GUGUGG,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF6 | 4 | 2997 | 0.023148148 | 0.0043447134 | 2.413564 | GUGUGG,UGUGGG,UGUGGU,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBMS3 | 1 | 1283 | 0.009259259 | 0.0018607779 | 2.314991 | UAUAGC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| TARDBP | 3 | 2654 | 0.018518519 | 0.0038476365 | 2.266924 | GAAUGA,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| HNRNPH3 | 9 | 7688 | 0.046296296 | 0.0111429292 | 2.054768 | AUGUGG,GGAAGG,GGAGGA,GGGAAG,GGGAAU,GGGAGG,GGGGAG,GUCGAG,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| RBM24 | 2 | 2357 | 0.013888889 | 0.0034172229 | 2.023035 | GUGUGG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF1 | 2 | 2391 | 0.013888889 | 0.0034664959 | 2.002381 | UGUGUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| PABPC5 | 2 | 2400 | 0.013888889 | 0.0034795387 | 1.996963 | AGAAAU,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SART3 | 2 | 2634 | 0.013888889 | 0.0038186524 | 1.862796 | AAAAAC,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| HNRNPH2 | 10 | 9711 | 0.050925926 | 0.0140746688 | 1.855299 | AUGUGG,CUGGGG,GGAAGG,GGAGGA,GGGAAG,GGGAAU,GGGAGG,GGGGAG,GUCGAG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| TRA2B | 7 | 7329 | 0.037037037 | 0.0106226650 | 1.801823 | AAAGAA,AAGAAU,AAGGAA,AGGAAA,GAAAGA,GAAGGA,GGAAGG | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| PUM2 | 1 | 1890 | 0.009259259 | 0.0027404447 | 1.756487 | UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPF | 10 | 10561 | 0.050925926 | 0.0153064921 | 1.734257 | AUGUGG,CUGGGG,GGAAGG,GGAGGA,GGGAAG,GGGAAU,GGGAGG,GGGGAG,UGGGAA,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 10 | 10717 | 0.050925926 | 0.0155325680 | 1.713104 | AUGUGG,CUGGGG,GGAAGG,GGAGGA,GGGAAG,GGGAAU,GGGAGG,GGGGAG,GUCGAG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| CSTF2 | 1 | 1967 | 0.009259259 | 0.0028520334 | 1.698906 | UGUGUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBMX | 3 | 4925 | 0.018518519 | 0.0071387787 | 1.375220 | AAGGAA,GAAGGA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| QKI | 1 | 2805 | 0.009259259 | 0.0040664663 | 1.187121 | AAUCAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| LIN28A | 2 | 4315 | 0.013888889 | 0.0062547643 | 1.150904 | CGGAGG,GGAGGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| PABPC1 | 2 | 4443 | 0.013888889 | 0.0064402624 | 1.108740 | AAAAAC,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| NELFE | 1 | 3028 | 0.009259259 | 0.0043896388 | 1.076795 | UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM4 | 1 | 3065 | 0.009259259 | 0.0044432593 | 1.059278 | UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| SRSF10 | 8 | 13860 | 0.041666667 | 0.0200874160 | 1.052602 | AAAGAA,AAAGAG,AAGAGA,AAGGAA,AGAGAC,GAAAGA,GAGAAA,GAGACC | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| ZRANB2 | 1 | 3173 | 0.009259259 | 0.0045997733 | 1.009334 | GUAAAG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
RBP on BSJ (Exon Only)
Plot
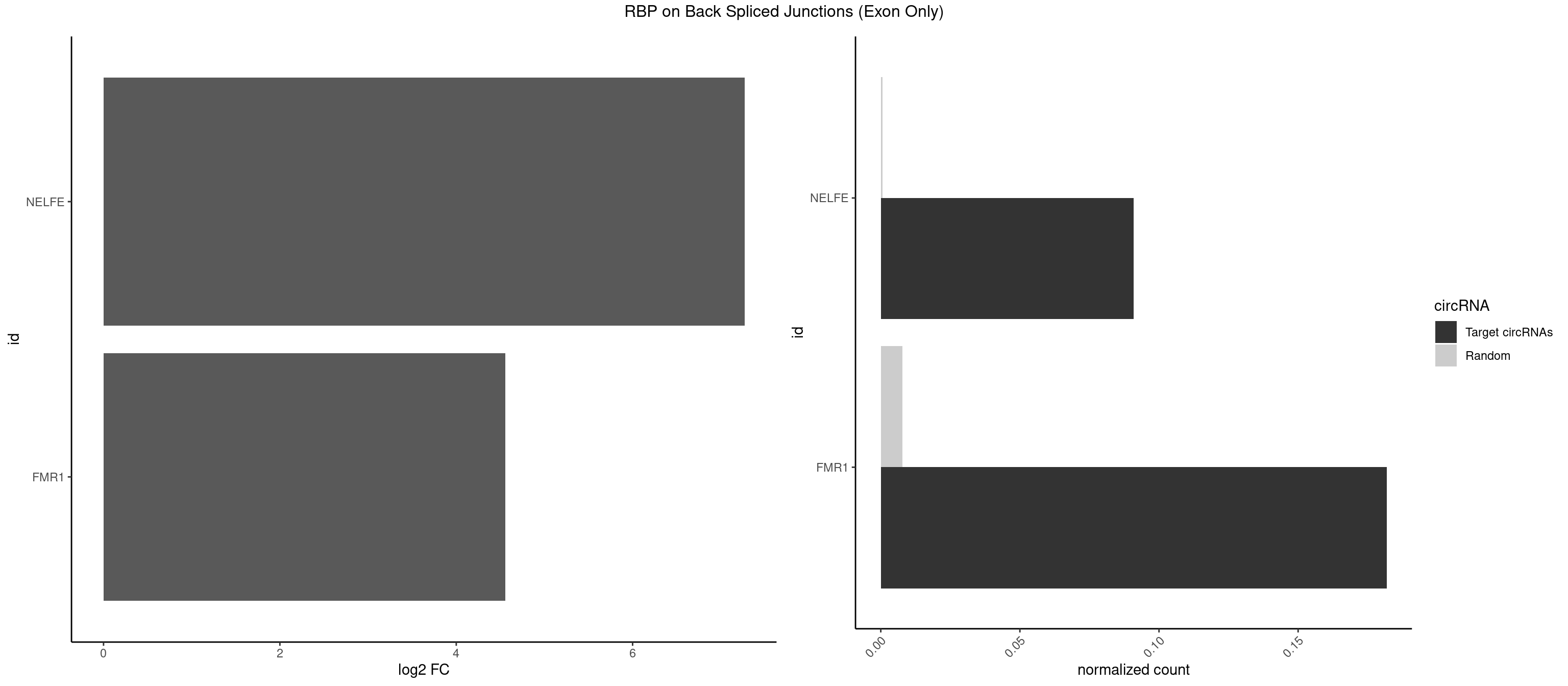
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NELFE | 1 | 8 | 0.09090909 | 0.0005894682 | 7.268867 | UGGCUA | CUCUGG,CUGGCU,CUGGUU,UCUGGC,UCUGGU |
| FMR1 | 3 | 117 | 0.18181818 | 0.0077285827 | 4.556149 | AGUGGC,GGCUAU,GUGGCU | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
RBP on BSJ (Exon and Intron)
Plot
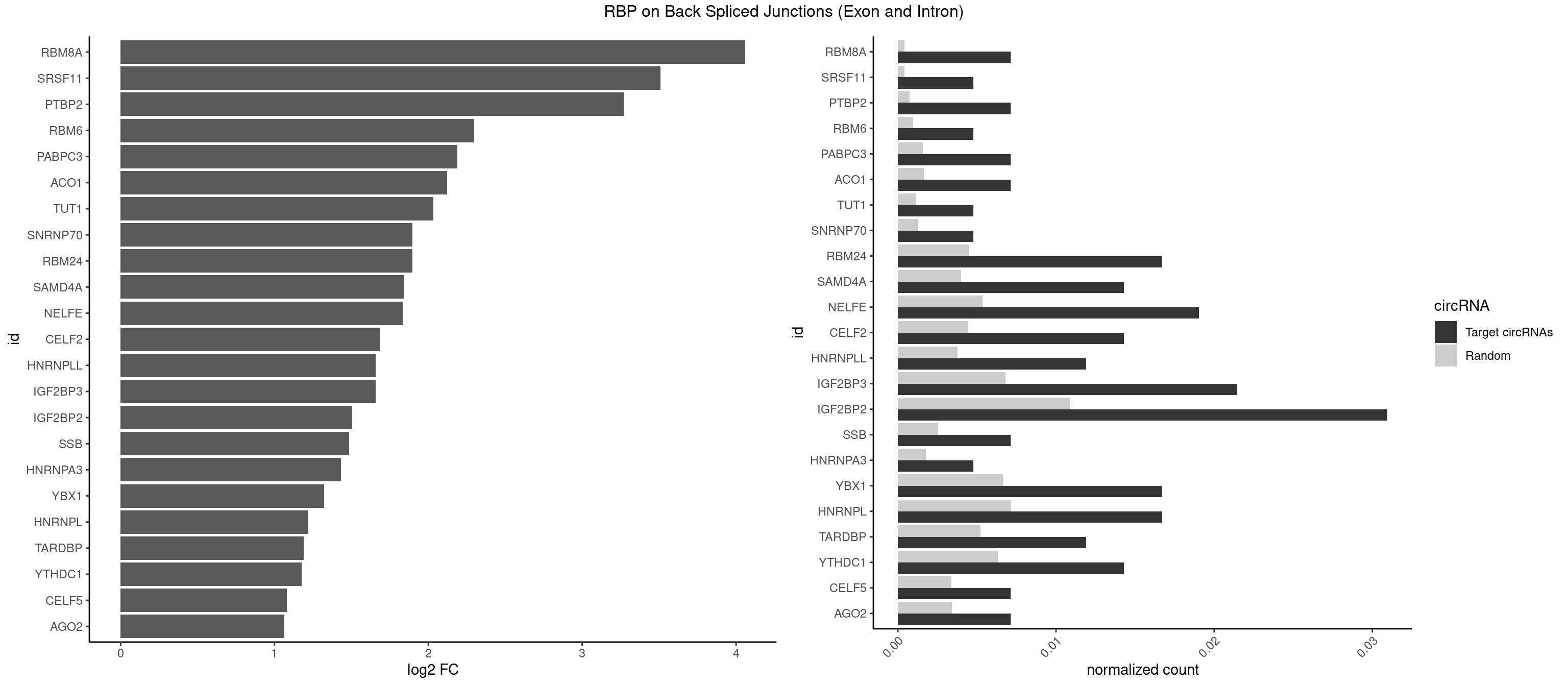
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM8A | 2 | 84 | 0.007142857 | 0.0004282547 | 4.059960 | AUGCGC,UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| PTBP2 | 2 | 146 | 0.007142857 | 0.0007406288 | 3.269679 | CUCUCU | CUCUCU |
| RBM6 | 1 | 191 | 0.004761905 | 0.0009673519 | 2.299426 | CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM24 | 6 | 888 | 0.016666667 | 0.0044790407 | 1.895704 | AGUGUG,GAGUGU,GUGUGA,GUGUGG,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| SAMD4A | 5 | 790 | 0.014285714 | 0.0039852882 | 1.841817 | CUGGAA,CUGGCA,CUGGCC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| NELFE | 7 | 1059 | 0.019047619 | 0.0053405885 | 1.834540 | CUCUCU,CUCUGG,CUGGCU,UCUCUG,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| CELF2 | 5 | 881 | 0.014285714 | 0.0044437727 | 1.684716 | AUGUGU,UAUGUG,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | ACAAAC,ACACCA,ACCACA,CACACC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| IGF2BP3 | 8 | 1348 | 0.021428571 | 0.0067966546 | 1.656639 | AAAACA,AAAAUC,AAAUCA,AAAUUC,AACACA,AAUUCA,ACAAAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| IGF2BP2 | 12 | 2163 | 0.030952381 | 0.0109028617 | 1.505344 | AAAACA,AAAAUC,AAAUCA,AAAUUC,AACACA,AAUUCA,ACAAAC,CCACAC,GAAAAC,GAACAC,GAAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| YBX1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | ACACCA,ACCACA,CACACC,CCACAA,CCACAC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| HNRNPL | 6 | 1419 | 0.016666667 | 0.0071543732 | 1.220068 | AAACAA,AACACA,ACACCA,ACCACA,CACAAA,CACACC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUGAAU,GUUGUG,UGUGUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GAAUGC,GCGUGC,GGAUAC,UCAUGC,UGGUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUGG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.