circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000131467:+:17:42838730:42839380
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000131467:+:17:42838730:42839380 | ENSG00000131467 | ENST00000545225 | + | 17 | 42838731 | 42839380 | 279 | GUCAAAAUGUGGGUACAGCUCCUGAUUCCCAGGAUAGAAGAUGGAAACAACUUUGGGGUGUCCAUUCAGGAGGAAACAGUUGCAGAGCUAAGAACUGUUGAGAGUGAAGCUGCAUCUUAUCUGGACCAGAUUUCUAGAUAUUAUAUUACAAGAGCCAAAUUGGUUUCUAAAAUAGCUAAAUAUCCCCAUGUGGAGGACUAUCGCCGCACCGUGACAGAGAUUGAUGAGAAAGAAUAUAUCAGCCUUCGGCUCAUCAUAUCAGAGCUGAGGAAUCAAUAUGUCAAAAUGUGGGUACAGCUCCUGAUUCCCAGGAUAGAAGAUGGAAACAA | circ |
| ENSG00000131467:+:17:42838730:42839380 | ENSG00000131467 | ENST00000545225 | + | 17 | 42838731 | 42839380 | 22 | GGAAUCAAUAUGUCAAAAUGUG | bsj |
| ENSG00000131467:+:17:42838730:42839380 | ENSG00000131467 | ENST00000545225 | + | 17 | 42838531 | 42838740 | 210 | UGGUCUCGAACUCCUGACCUCAGGUGAUCCACCCGCUUCGGGGUCCCAAAUUGCUGGGAUUACCGGCAUGAGCCACCAUGCCUGGCCAGACUAGGUAUUUUGACUUCUGUGUUCCUGGCAUCUGGCUCCAGCCGUCUGAAUUGUGUACUUCAUGGCUUCAGCCUUCAGGCAAAGGUCCUCAUAUAUGUUUUGACCUCCAGGUCAAAAUGU | ie_up |
| ENSG00000131467:+:17:42838730:42839380 | ENSG00000131467 | ENST00000545225 | + | 17 | 42839371 | 42839580 | 210 | GAAUCAAUAUGUGAGUAAUCUCAGUCCUUGUCCAUAGUUGCUGUGGCUUCUCUUAGUUCUGUUGAGAGUAUUGUUAAAAUUCUCUGAAGGGCUGAGAUGAGGGUCUGAGGUAAAAUGAAUAGACCAAAUUCUAUUCAUCACCUGAGUGUUGAGAACCAUGAAGUCAUCUAGCAGGGUAUCUCUCUUUUUUCUACCUGAGUGGAGGGGUGU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
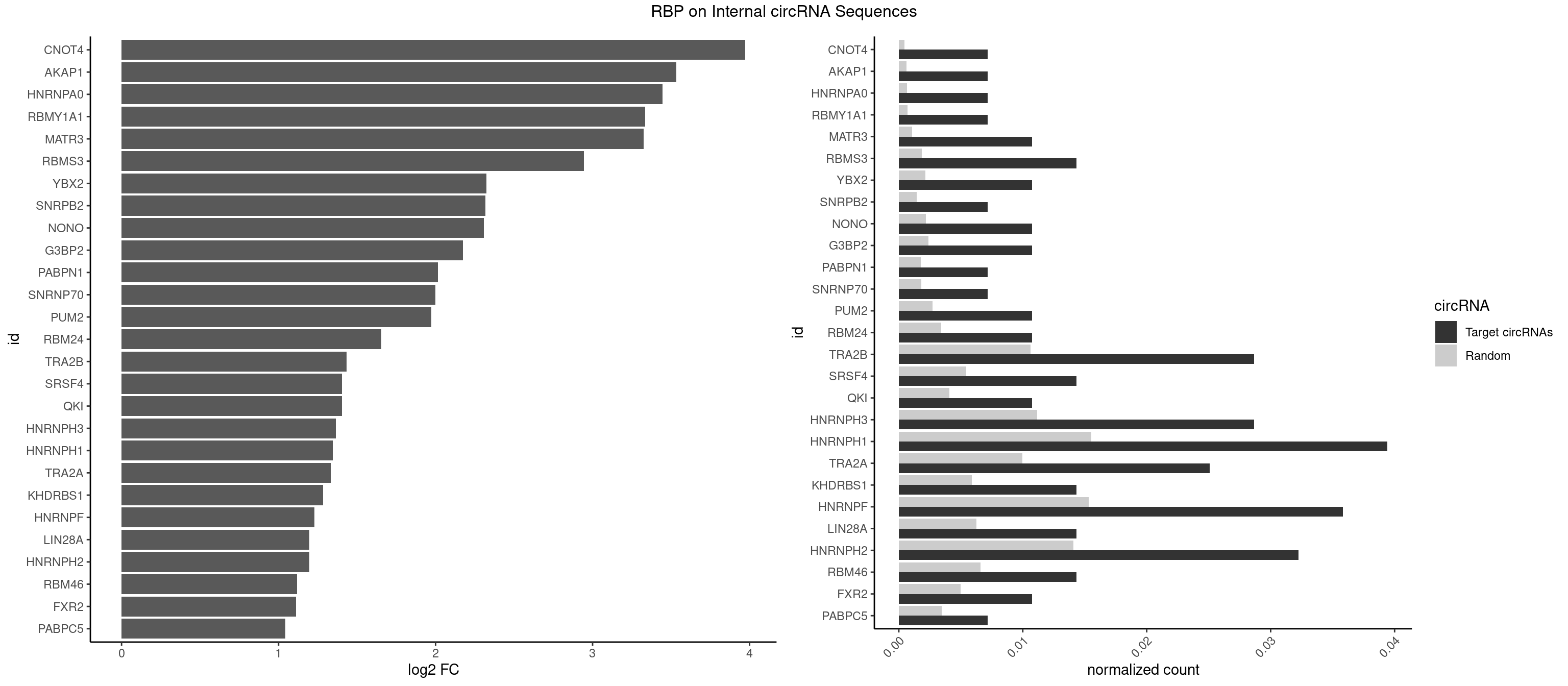
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 314 | 0.007168459 | 0.0004564992 | 3.972979 | GACAGA | GACAGA |
| AKAP1 | 1 | 426 | 0.007168459 | 0.0006188101 | 3.534094 | AUAUAU | AUAUAU,UAUAUA |
| HNRNPA0 | 1 | 453 | 0.007168459 | 0.0006579386 | 3.445638 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMY1A1 | 1 | 489 | 0.007168459 | 0.0007101099 | 3.335549 | ACAAGA | ACAAGA,CAAGAC |
| MATR3 | 2 | 739 | 0.010752688 | 0.0010724109 | 3.325768 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMS3 | 3 | 1283 | 0.014336918 | 0.0018607779 | 2.945757 | AAUAUA,AUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| YBX2 | 2 | 1480 | 0.010752688 | 0.0021462711 | 2.324793 | AACAAC,ACAACU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SNRPB2 | 1 | 991 | 0.007168459 | 0.0014376103 | 2.317990 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| NONO | 2 | 1498 | 0.010752688 | 0.0021723567 | 2.307364 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| G3BP2 | 2 | 1644 | 0.010752688 | 0.0023839405 | 2.173277 | AGGAUA,GGAUAG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| PABPN1 | 1 | 1222 | 0.007168459 | 0.0017723764 | 2.015978 | AGAAGA | AAAAGA,AGAAGA |
| SNRNP70 | 1 | 1237 | 0.007168459 | 0.0017941145 | 1.998391 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PUM2 | 2 | 1890 | 0.010752688 | 0.0027404447 | 1.972215 | UAAAUA,UAGAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| RBM24 | 2 | 2357 | 0.010752688 | 0.0034172229 | 1.653801 | AGAGUG,GAGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| TRA2B | 7 | 7329 | 0.028673835 | 0.0106226650 | 1.432589 | AAAGAA,AAGAAC,AAGAAU,AGAAGA,AGGAAA,GAAAGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SRSF4 | 3 | 3740 | 0.014336918 | 0.0054214720 | 1.402978 | AGAAGA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| QKI | 2 | 2805 | 0.010752688 | 0.0040664663 | 1.402850 | AUCAUA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPH3 | 7 | 7688 | 0.028673835 | 0.0111429292 | 1.363606 | AAUGUG,AUGUGG,GGAGGA,GGGUGU,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 10 | 10717 | 0.039426523 | 0.0155325680 | 1.343870 | AAUGUG,AUGUGG,GAGGAA,GGAGGA,GGGUGU,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| TRA2A | 6 | 6871 | 0.025089606 | 0.0099589296 | 1.333027 | AAAGAA,AGAAAG,AGAAGA,GAAAGA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| KHDRBS1 | 3 | 4064 | 0.014336918 | 0.0058910141 | 1.283147 | CAAAAU,CUAAAA,CUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| HNRNPF | 9 | 10561 | 0.035842294 | 0.0153064921 | 1.227519 | AAUGUG,AUGUGG,GAGGAA,GGAGGA,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| LIN28A | 3 | 4315 | 0.014336918 | 0.0062547643 | 1.196707 | GGAGGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPH2 | 8 | 9711 | 0.032258065 | 0.0140746688 | 1.196559 | AAUGUG,AUGUGG,GGAGGA,GGGUGU,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| RBM46 | 3 | 4554 | 0.014336918 | 0.0066011240 | 1.118951 | AAUCAA,AUCAAU,AUCAUA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| FXR2 | 2 | 3434 | 0.010752688 | 0.0049780156 | 1.111055 | GACAGA,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| PABPC5 | 1 | 2400 | 0.007168459 | 0.0034795387 | 1.042767 | AGAAAG | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
RBP on BSJ (Exon Only)
Plot
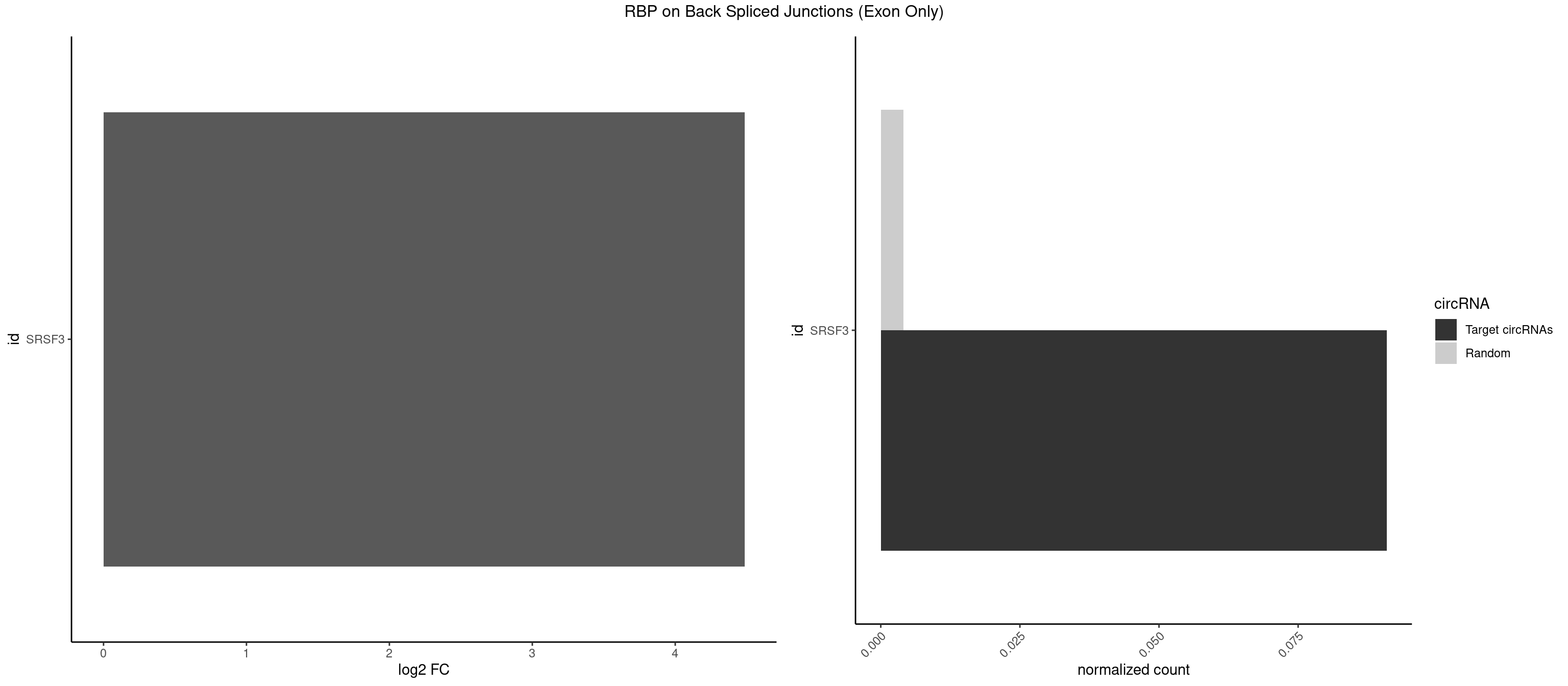
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF3 | 1 | 61 | 0.09090909 | 0.004060781 | 4.484596 | UGUCAA | AACGAU,ACCACC,ACUACG,AGAGAU,CACAAC,CACAUC,CACCAC,CAGAGA,CAUCAC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CUACAG,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUUAU,GACUUC,GAGAUU,UACAGC,UCAGAG,UCUCCA,UCUUCA,UCUUCC,UGUCAA,UUCGAC,UUCUCC,UUCUUC |
RBP on BSJ (Exon and Intron)
Plot
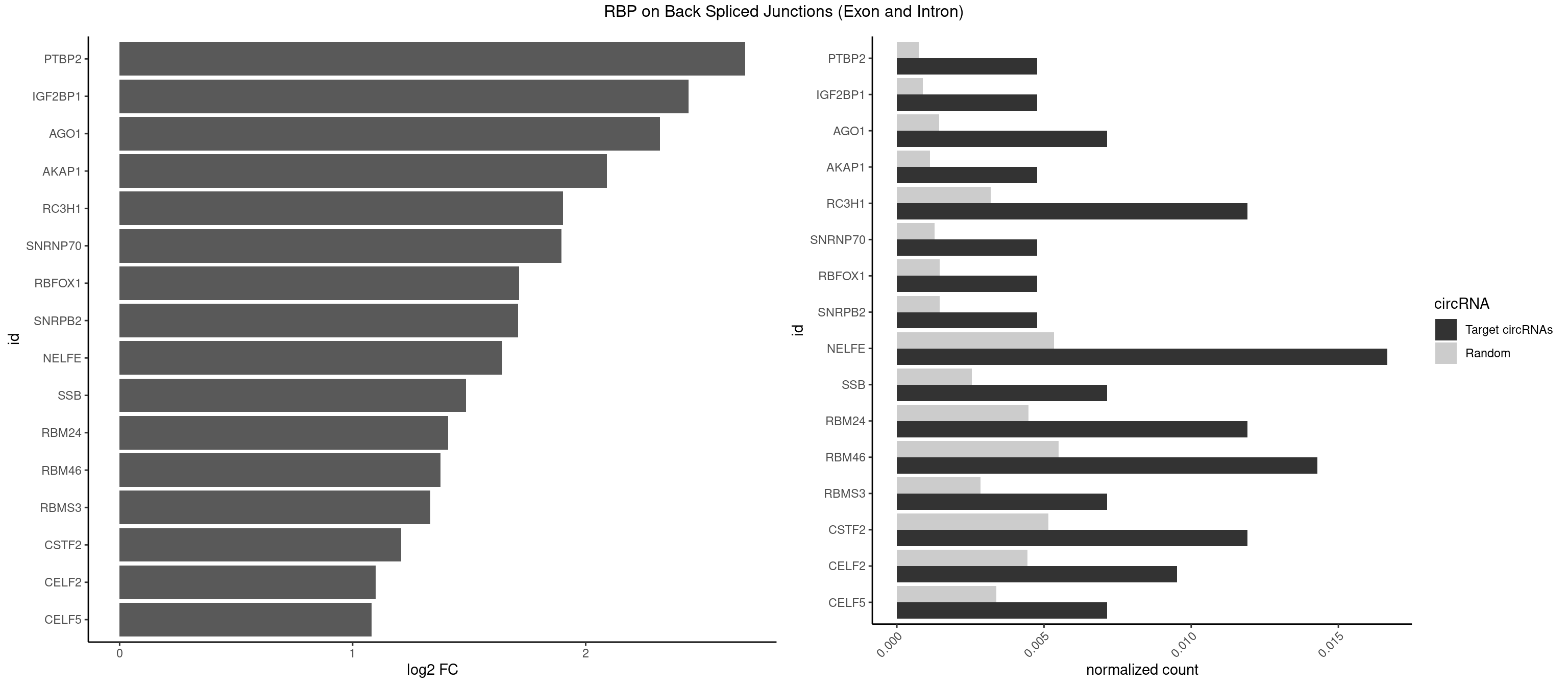
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | CACCCG | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | GAGGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| RC3H1 | 4 | 631 | 0.011904762 | 0.0031841999 | 1.902536 | CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| NELFE | 6 | 1059 | 0.016666667 | 0.0053405885 | 1.641895 | CUCUCU,CUGGCU,GGUCUC,UCUCUC,UCUCUG,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | GAGUGG,GAGUGU,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| RBM46 | 5 | 1091 | 0.014285714 | 0.0055018138 | 1.376594 | AAUCAA,AAUGAA,AUCAAU,AUGAAG,AUGAAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | AUAUAU,CAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUUG,GUUUUG,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | UAUGUG,UAUGUU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.