circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000111358:+:12:123645454:123652735
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000111358:+:12:123645454:123652735 | ENSG00000111358 | MSTRG.8256.3 | + | 12 | 123645455 | 123652735 | 393 | UUCACUUUAUCCAAAUGCAUAGAUGCCGUGAUGGUGCUGGGAAAUUCGCAUUUAUUCAUGAAUCGUUCCAACAAACUUGCUGUGAUAGCAAGUCACAUUCAAGAAAGCCGAUUCUUAUAUCCUGGAAAGAAUGGCAGACUUGGAGACUUCUUCGGAGACCCUGGCAACCCUCCUGAAUUUAAUCCCUCUGGGAGUAAAGAUGGAAAAUACGAACUUUUAACCUCAGCAAAUGAAGUUAUUGUUGAAGAGAUUAAAGAUCUAAUGACCAAAAGUGACAUAAAGGGUCAACAUACAGAAACUUUGCUGGCAGGAUCCCUGGCCAAAGCCCUUUGCUACAUUCAUAGAAUGAACAAGGAAGUUAAAGACAAUCAGGAAAUGAAAUCAAGGAUAUUGUUCACUUUAUCCAAAUGCAUAGAUGCCGUGAUGGUGCUGGGAAAUUCGCA | circ |
| ENSG00000111358:+:12:123645454:123652735 | ENSG00000111358 | MSTRG.8256.3 | + | 12 | 123645455 | 123652735 | 22 | CAAGGAUAUUGUUCACUUUAUC | bsj |
| ENSG00000111358:+:12:123645454:123652735 | ENSG00000111358 | MSTRG.8256.3 | + | 12 | 123645255 | 123645464 | 210 | AGCUAAGGUUUCAUUAUGUUGCCUAGGCUGGUCUCAAACUCCUGAACUCAAGCAGUCCUCCCACCUUGGCCUCUCAGGGUGCUGGGAUUACAGGCUUGAGCCACUGCACCCAGCCUAAACGACAUCUUAAUUAUGUCAAGACCUGACAUGGUUAUUUGGAUAGCUAAUUUGUUUUUCUAAUGUCUUUUUUUUUCCAACAGUUCACUUUAU | ie_up |
| ENSG00000111358:+:12:123645454:123652735 | ENSG00000111358 | MSTRG.8256.3 | + | 12 | 123652726 | 123652935 | 210 | AAGGAUAUUGGUAAGAUAAAAAAAUCAUUACUUUUUUAUAUGACAACUAUAAUUAGAAAACAUAACGGGAGACCAGGUGUGGUGGCUCACACCUGUUAUCCCAGCACUUUGGGAGGCCGAGGUGGGCAGAUCACUUGAGGUCAGGAGUUCGGGACCAGCCUGGCCAACAUGGUGAAACCCCGUCUCUACUCAAAAUACAAAAAUUAGCUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
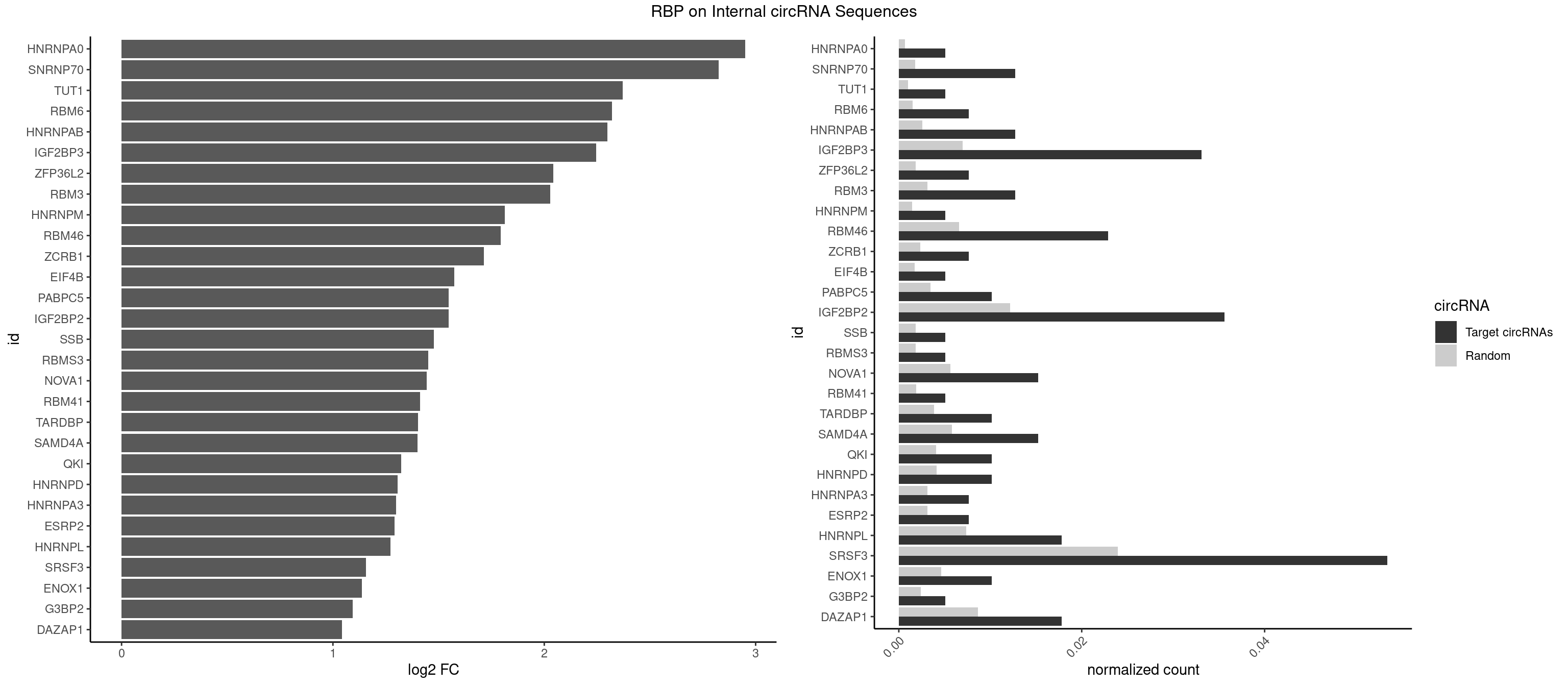
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 1 | 453 | 0.005089059 | 0.0006579386 | 2.951374 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| SNRNP70 | 4 | 1237 | 0.012722646 | 0.0017941145 | 2.826055 | AAUCAA,AUCAAG,AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| TUT1 | 1 | 678 | 0.005089059 | 0.0009840095 | 2.370655 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM6 | 2 | 1054 | 0.007633588 | 0.0015289102 | 2.319858 | AUCCAA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPAB | 4 | 1782 | 0.012722646 | 0.0025839306 | 2.299760 | AAAGAC,AAGACA,AGACAA,AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| IGF2BP3 | 12 | 4815 | 0.033078880 | 0.0069793662 | 2.244742 | AAAUAC,AAAUCA,AAAUUC,ACAAAC,ACAAUC,ACAUAC,ACAUUC,CAAUCA,CAUACA,CAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| ZFP36L2 | 2 | 1277 | 0.007633588 | 0.0018520827 | 2.043213 | AUUUAU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM3 | 4 | 2152 | 0.012722646 | 0.0031201361 | 2.027718 | AAUACG,AUACGA,GAAACU,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPM | 1 | 999 | 0.005089059 | 0.0014492040 | 1.812138 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| RBM46 | 8 | 4554 | 0.022900763 | 0.0066011240 | 1.794612 | AAUCAA,AAUGAA,AUCAAG,AUGAAA,AUGAAG,AUGAAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ZCRB1 | 2 | 1605 | 0.007633588 | 0.0023274215 | 1.713629 | AUUUAA,GAUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| EIF4B | 1 | 1179 | 0.005089059 | 0.0017100607 | 1.573351 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPC5 | 3 | 2400 | 0.010178117 | 0.0034795387 | 1.548503 | AGAAAG,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP2 | 13 | 8408 | 0.035623410 | 0.0121863560 | 1.547559 | AAAUAC,AAAUCA,AAAUUC,ACAAAC,ACAAUC,ACAUAC,ACAUUC,CAAUCA,CAUACA,CAUUCA,GAAAUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| SSB | 1 | 1260 | 0.005089059 | 0.0018274462 | 1.477570 | UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBMS3 | 1 | 1283 | 0.005089059 | 0.0018607779 | 1.451493 | UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| NOVA1 | 5 | 3872 | 0.015267176 | 0.0056127669 | 1.443649 | AGUCAC,AUUCAU,CAUUCA | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| RBM41 | 1 | 1318 | 0.005089059 | 0.0019115000 | 1.412694 | UACAUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| TARDBP | 3 | 2654 | 0.010178117 | 0.0038476365 | 1.403426 | GAAUGA,GAAUGG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SAMD4A | 5 | 3992 | 0.015267176 | 0.0057866714 | 1.399628 | CUGGAA,CUGGCA,CUGGCC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| QKI | 3 | 2805 | 0.010178117 | 0.0040664663 | 1.323623 | AUCUAA,UAACCU,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPD | 3 | 2837 | 0.010178117 | 0.0041128408 | 1.307264 | AAUUUA,AUUUAU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| HNRNPA3 | 2 | 2140 | 0.007633588 | 0.0031027457 | 1.298816 | CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 2 | 2150 | 0.007633588 | 0.0031172377 | 1.292093 | GGGAAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPL | 6 | 5085 | 0.017811705 | 0.0073706513 | 1.272962 | AAAUAC,AACAAA,ACAUAA,ACAUAC,CAUAAA,CAUACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| SRSF3 | 20 | 16536 | 0.053435115 | 0.0239654858 | 1.156830 | AACUUU,ACAUUC,ACUUCU,ACUUUA,AGAGAU,AUCGUU,AUUCAU,CAUUCA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GUCAAC,UCGUUC,UCUUCG,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| ENOX1 | 3 | 3195 | 0.010178117 | 0.0046316558 | 1.135871 | AAGACA,AUACAG,CAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| G3BP2 | 1 | 1644 | 0.005089059 | 0.0023839405 | 1.094051 | AGGAUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| DAZAP1 | 6 | 5964 | 0.017811705 | 0.0086445016 | 1.042971 | AAUUUA,AGGAAA,AGGAAG,AGGAUA,AGUAAA,AGUUAA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
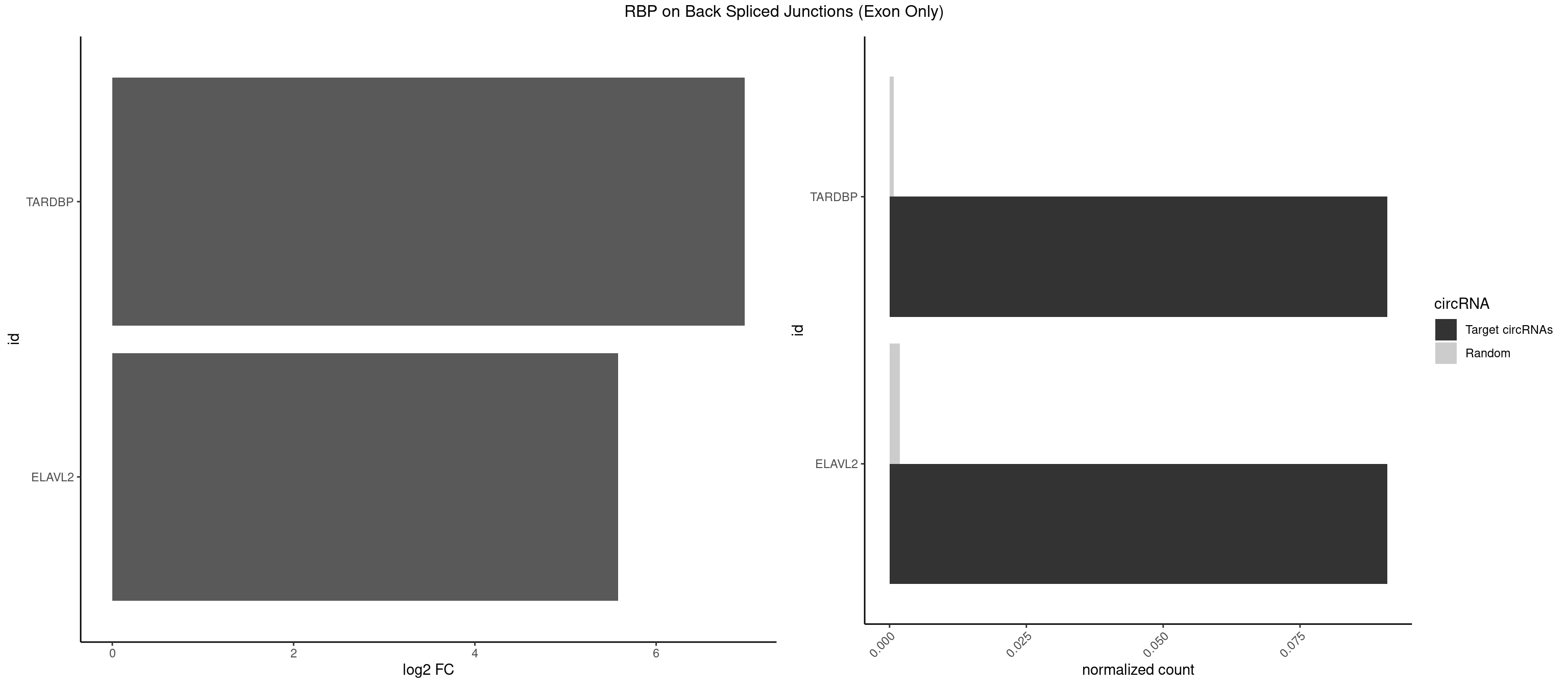
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TARDBP | 1 | 10 | 0.09090909 | 0.0007204611 | 6.979360 | UUGUUC | GAAUGA,GAAUGU,GUUUUG,UGUGUG,UUGUGC,UUGUUC |
| ELAVL2 | 1 | 28 | 0.09090909 | 0.0018993974 | 5.580811 | UAUUGU | AAUUUA,AAUUUU,AUAUUU,AUUUAA,AUUUUG,CUUUCU,GAUUUU,GUAUUG,GUUUUU,UACUUU,UAGUUA,UAUAUA,UAUAUU,UAUGUU,UAUUGA,UAUUUU,UGUAUU,UUAAGU,UUACUU,UUAGUU,UUAUUG,UUUACU,UUUAGU,UUUCUU,UUUUAG |
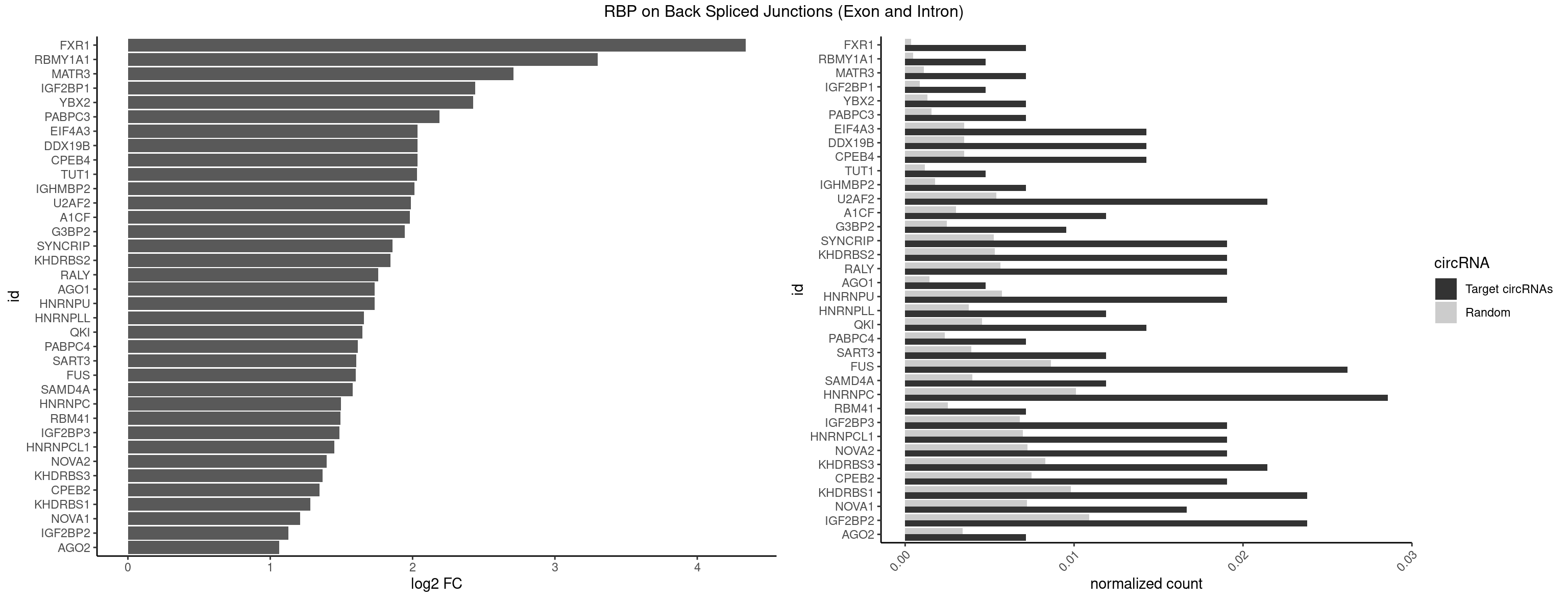
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 2 | 69 | 0.007142857 | 0.0003526804 | 4.340068 | ACGACA,AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | ACAACU,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| CPEB4 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| DDX19B | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| IGHMBP2 | 2 | 350 | 0.007142857 | 0.0017684401 | 2.014024 | AAAAAA | AAAAAA |
| U2AF2 | 8 | 1071 | 0.021428571 | 0.0054010480 | 1.988224 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUA,GGAUAG,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SYNCRIP | 7 | 1041 | 0.019047619 | 0.0052498992 | 1.859249 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 7 | 1051 | 0.019047619 | 0.0053002821 | 1.845470 | AUAAAA,GAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RALY | 7 | 1117 | 0.019047619 | 0.0056328094 | 1.757684 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| HNRNPU | 7 | 1136 | 0.019047619 | 0.0057285369 | 1.733372 | AAAAAA,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | ACGACA,ACUGCA,CACACC,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | AAUCAU,CUACUC,UACUCA,UCUAAU,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA | AAAAAA,AAAAAG |
| SART3 | 4 | 777 | 0.011904762 | 0.0039197904 | 1.602690 | AAAAAA,AGAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| FUS | 10 | 1711 | 0.026190476 | 0.0086255542 | 1.602353 | AAAAAA,GGGUGC,UGGUGA,UGGUGG,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| SAMD4A | 4 | 790 | 0.011904762 | 0.0039852882 | 1.578783 | CGGGAC,CUGGCC,CUGGUC,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| HNRNPC | 11 | 2006 | 0.028571429 | 0.0101118501 | 1.498526 | CUUUUU,GGAUAU,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| IGF2BP3 | 7 | 1348 | 0.019047619 | 0.0067966546 | 1.486714 | AAAACA,AAAAUC,AAACUC,AAAUAC,AAAUCA,AACUCA,AAUACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| HNRNPCL1 | 7 | 1381 | 0.019047619 | 0.0069629182 | 1.451847 | CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| NOVA2 | 7 | 1434 | 0.019047619 | 0.0072299476 | 1.397554 | AGAUCA,GAUCAC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS3 | 8 | 1646 | 0.021428571 | 0.0082980653 | 1.368689 | AGAUAA,AUAAAA,GAUAAA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| CPEB2 | 7 | 1487 | 0.019047619 | 0.0074969770 | 1.345230 | CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| KHDRBS1 | 9 | 1945 | 0.023809524 | 0.0098045143 | 1.280021 | AUAAAA,CAAAAU,GAAAAC,UAAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| NOVA1 | 6 | 1428 | 0.016666667 | 0.0071997179 | 1.210953 | UUCAUU,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| IGF2BP2 | 9 | 2163 | 0.023809524 | 0.0109028617 | 1.126832 | AAAACA,AAAAUC,AAACUC,AAAUAC,AAAUCA,AACUCA,AAUACA,GAAAAC,GAACUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.