circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000171045:-:8:142330900:142354763
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000171045:-:8:142330900:142354763 | ENSG00000171045 | MSTRG.32164.6 | - | 8 | 142330901 | 142354763 | 932 | CUCAGUGUGGAGGCUGCUGGCCCUGCCAUCUGUAAGAAGAUGUCAUACGGAUCCAUCGCCCGUGGAGGUGGCCUGGGGAGCCGUGGCCCUUUCGGGGGACCUUCGAGACAAGGCUGUCAGCCCCUAGAGUGCGCUAGAUGUUGGACCGAGUAUGGAAUCCGCCAUUUCCCCUGCCCGUCGCCAGAGAGCAAGCUGCAGAACCGCUGUGUGGGGAAGGACGGGGAAGGUGAUCUGGGGCCAGCAGGCACGCCUAUUGUCCCAAGGGCCAGGAAGCGAGGGCCUGGGGUUGCCCCUGAAGGCAGCCGGAUGCCGGAGCCCACCUCAUCACCCACCAUUGGCCCGAGGAAGGACUCGGCUGCUGGGCCCCAUGGCCGGAUGGCGGGGCCCAGCACUACCCGGGCCAAGAAGAGGAAGCCCAACUUCUGCCCGCAGGAGACCGAGGUGCUGGUGUCCAAGGUGAGCAAGCACCACCAGCUGCUGUUUGGCACGGGGCUGCUGAAGGCCGAGCCCACUCGCAGGUACCGCGUGUGGAGCCGCAUCCUGCAGGCCGUGAAUGCGCUGGGCUACUGUCGCCGCGACGUUGUGGACCUGAAGCACAAGUGGCGGGACCUACGAGCCGUCGUGCGGCGCAAGCUGGGCGACCUCCGGAAGGCGGCCCAUGGCCCCAGCCCUGGUUCCGGCAAGCCCCAGGCCCUGGCUCUCACGCCCGUGGAGCAGGUGGUGGCCAAGACCUUCUCUUGCCAGGCCCUGCCCUCCGAGGGCUUCAGUCUGGAGCCGCCCAGAGCCACCCAGGUCGAUCCGUGCAACCUCCAGGAGCUGUUCCAGGAGAUGUCGGCCAACGUCUUCCGAAUCAACUCCAGUGUGACCUCCUUGGAGCGGAGCCUUCAGUCCUUAGGGACACCGAGUGACACGCAGGAGCUUCGGGACAGCCUCUCAGUGUGGAGGCUGCUGGCCCUGCCAUCUGUAAGAAGAUGUCAUACGG | circ |
| ENSG00000171045:-:8:142330900:142354763 | ENSG00000171045 | MSTRG.32164.6 | - | 8 | 142330901 | 142354763 | 22 | CGGGACAGCCUCUCAGUGUGGA | bsj |
| ENSG00000171045:-:8:142330900:142354763 | ENSG00000171045 | MSTRG.32164.6 | - | 8 | 142354754 | 142354963 | 210 | CAAGAGGCCACCUUGAAGGGGCUCUGUGGGGGGGUUGCUGUUUGGGGGCUGGUGAGAAGCCUGGAGUAGCCUAGGGAAUGGGUACAAUGGAGCCAGGGUCUUGGGGCUGGGGAGGGGCCGGCGGCCCCUGUGCACCCUGAAUUGGAGCUUUAAUCCCCAUGUCUUCCCCCUCUUCCCCCUGCUUUUCGUGUUUCCUCCAGCUCAGUGUGG | ie_up |
| ENSG00000171045:-:8:142330900:142354763 | ENSG00000171045 | MSTRG.32164.6 | - | 8 | 142330701 | 142330910 | 210 | GGGACAGCCUGUGAGUGUGUGGGCCAUGGUAUCUCUUUGCCUGGGCGUGGUGCACAUUGGGGGGCAGGGGCGGGAGUGUGGUCCUCCUGUGCACACGGGCUUUGUCCACACAGCUGCUUGCGCACAGCUGCCUGGACACACAUGUGCGCACAUAUGCACACACGCCUGCGGAUAAGCCUAGUCAGUGCUGUGAACCUGAGACUGGGCCCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
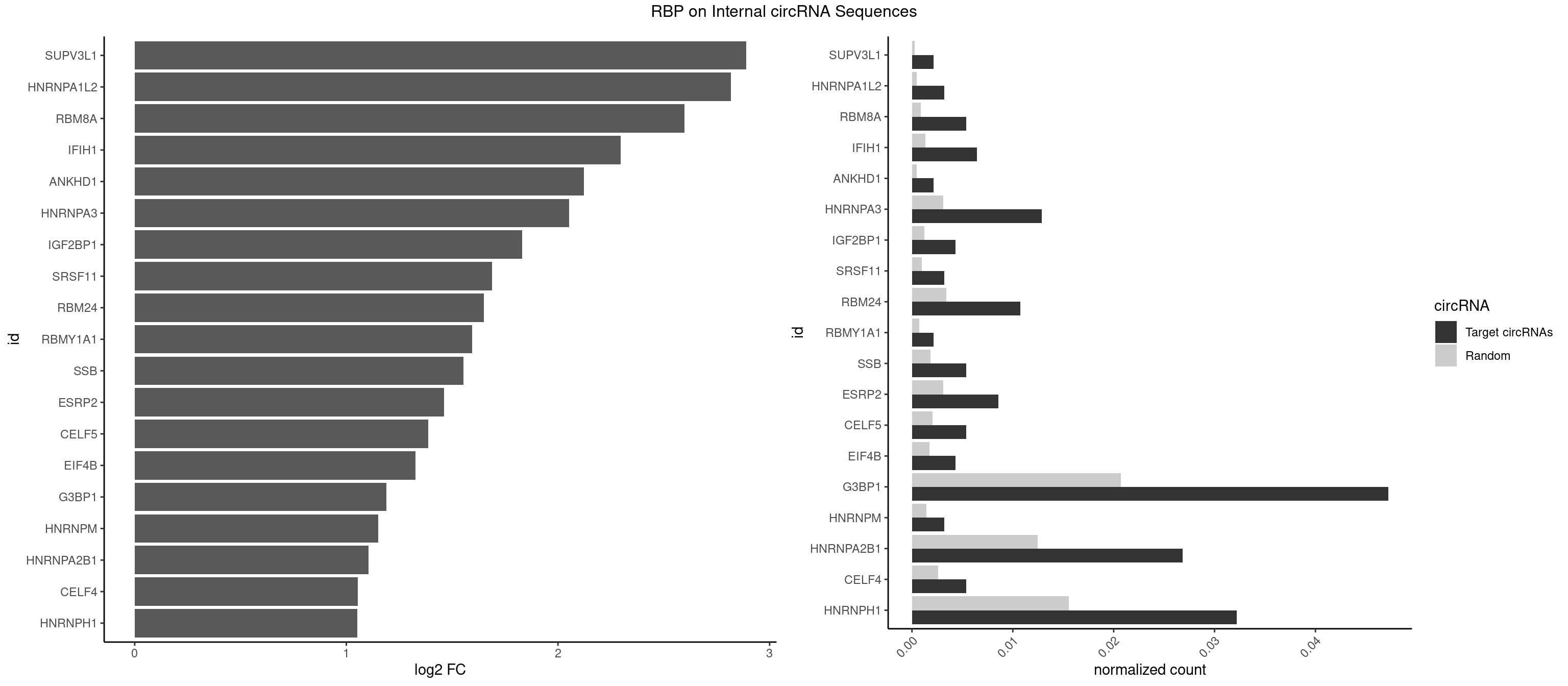
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 1 | 199 | 0.002145923 | 0.0002898408 | 2.888266 | CCGCCC | CCGCCC |
| HNRNPA1L2 | 2 | 314 | 0.003218884 | 0.0004564992 | 2.817876 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBM8A | 4 | 611 | 0.005364807 | 0.0008869128 | 2.596662 | AUGCGC,GUGCGC,UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| IFIH1 | 5 | 904 | 0.006437768 | 0.0013115296 | 2.295310 | GCCGCG,GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ANKHD1 | 1 | 339 | 0.002145923 | 0.0004927293 | 2.122731 | GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| HNRNPA3 | 11 | 2140 | 0.012875536 | 0.0031027457 | 2.053015 | AGGAGC,CCAAGG,GCCAAG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| IGF2BP1 | 3 | 831 | 0.004291845 | 0.0012057377 | 1.831682 | AAGCAC,AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SRSF11 | 2 | 688 | 0.003218884 | 0.0009985015 | 1.688724 | AAGAAG | AAGAAG |
| RBM24 | 9 | 2357 | 0.010729614 | 0.0034172229 | 1.650702 | AGAGUG,AGUGUG,GAGUGA,GUGUGA,GUGUGG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| RBMY1A1 | 1 | 489 | 0.002145923 | 0.0007101099 | 1.595484 | CAAGAC | ACAAGA,CAAGAC |
| SSB | 4 | 1260 | 0.005364807 | 0.0018274462 | 1.553697 | CUGUUU,GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ESRP2 | 7 | 2150 | 0.008583691 | 0.0031172377 | 1.461330 | GGGAAG,GGGGAA,GGGGAG,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| CELF5 | 4 | 1415 | 0.005364807 | 0.0020520728 | 1.386444 | GUGUGG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| EIF4B | 3 | 1179 | 0.004291845 | 0.0017100607 | 1.327551 | CUUGGA,GUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| G3BP1 | 43 | 14282 | 0.047210300 | 0.0206989801 | 1.189542 | ACACGC,ACCCAC,ACGCAG,ACGCCC,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CCACCC,CCAGGC,CCAUCG,CCCACC,CCCAGG,CCCCAG,CCCCUA,CCCGCA,CCCUAG,CCCUCC,CCGCAG,CCGCCC,CCGGCA,CCUACG,CCUCCG,CGGCCC,CUCGGC,UCCGCC,UCGGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| HNRNPM | 2 | 999 | 0.003218884 | 0.0014492040 | 1.151300 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| HNRNPA2B1 | 24 | 8607 | 0.026824034 | 0.0124747476 | 1.104516 | AAGAAG,AGGAGC,CAAGAA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GGAGCC,GGGGCC,UAGGGA,UUAGGG | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| CELF4 | 4 | 1782 | 0.005364807 | 0.0025839306 | 1.053959 | GUGUGG,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPH1 | 29 | 10717 | 0.032188841 | 0.0155325680 | 1.051264 | AAGGCG,AAGGUG,AGGAAG,CGAGGG,CGGGGG,CUGGGG,GAAGGU,GAGGAA,GGAAGG,GGGAAG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGUCGA,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
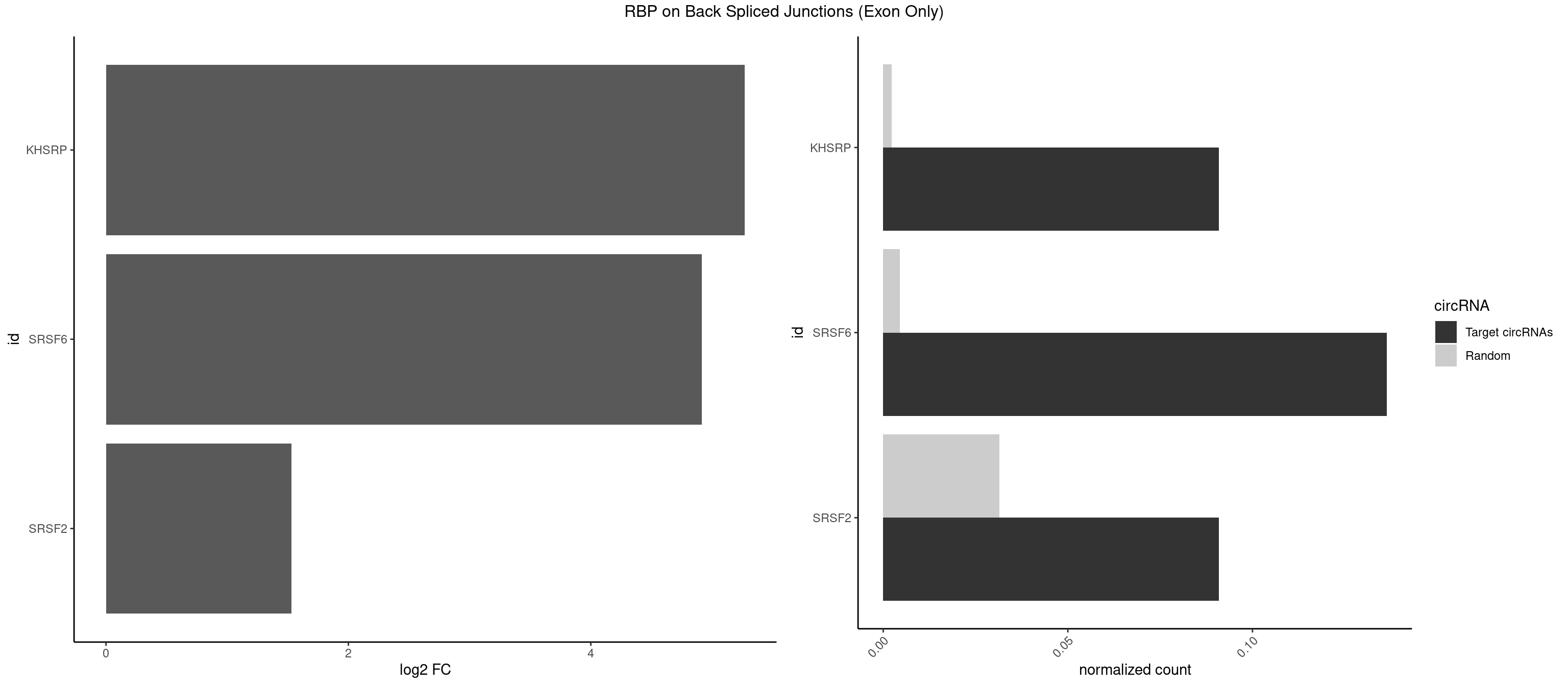
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| KHSRP | 1 | 35 | 0.09090909 | 0.002357873 | 5.268867 | AGCCUC | ACCCUC,AGCCUC,AUAUUU,AUGUAU,AUGUGU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CGGCUU,CUCCCU,CUGCCU,CUGCUU,GCCCUC,GCCUCC,GCCUUC,GGCCUC,UAGGUU,UAUAUU,UAUUUU,UCCCUC,UGCAUG,UGUAUA,UGUGUG,UUAUUA |
| SRSF6 | 2 | 68 | 0.13636364 | 0.004519256 | 4.915230 | CCUCUC,UCUCAG | AACCUG,ACCGUC,ACCUGG,AGAAGA,AGCGGA,AGGAAG,AUCCUG,CAACCU,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CCUGGC,CUACAG,CUCAGG,CUCUGG,CUUCUG,GAAGAA,GAGGAA,GCACCU,GCAGCA,GGAAGA,UACAGG,UACUGG,UCCUGG,UGCGGC,UGUGGA,UUACUG,UUCAGG,UUCUGG,UUUCAG |
| SRSF2 | 1 | 479 | 0.09090909 | 0.031438302 | 1.531901 | AGCCUC | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
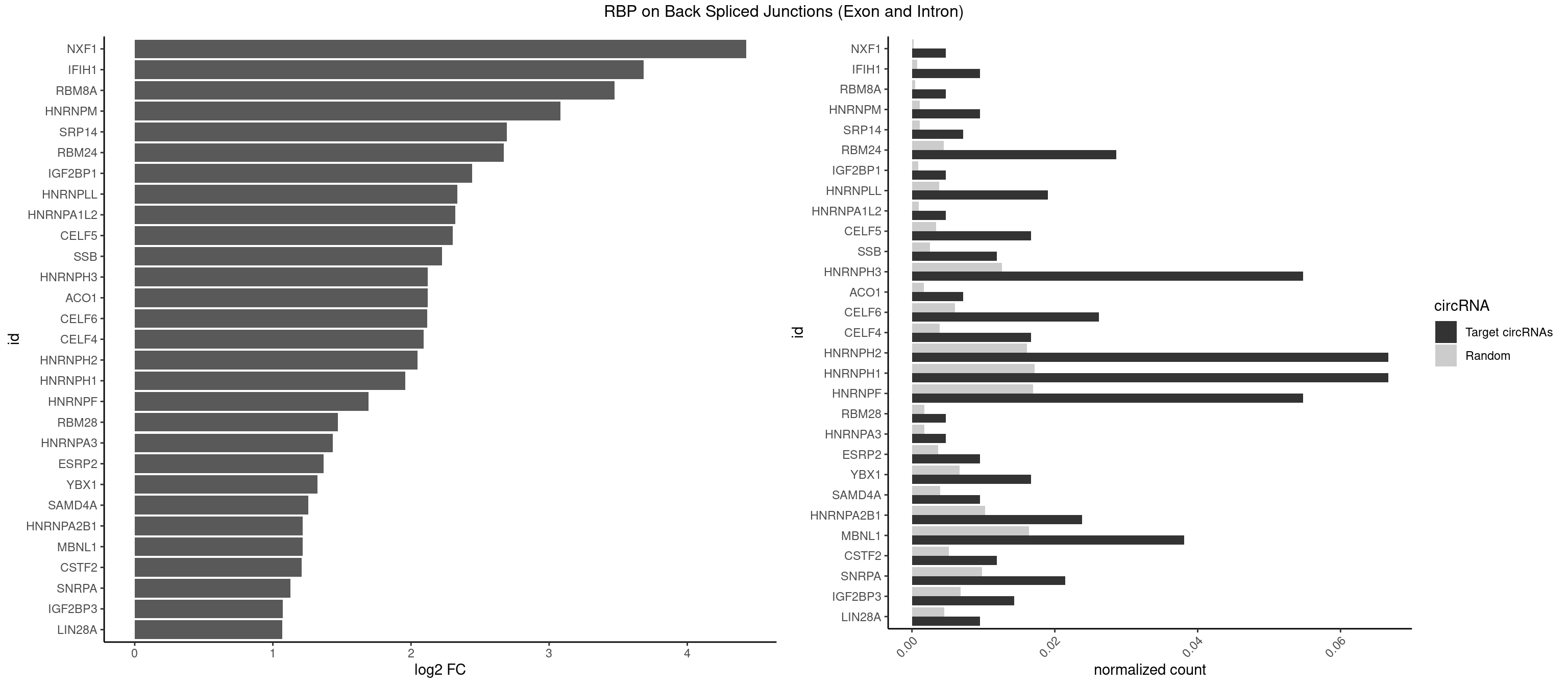
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| IFIH1 | 3 | 146 | 0.009523810 | 0.0007406288 | 3.684716 | GCGGAU,GGCCCU,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM8A | 1 | 84 | 0.004761905 | 0.0004282547 | 3.474998 | GUGCGC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| HNRNPM | 3 | 222 | 0.009523810 | 0.0011235389 | 3.083489 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM24 | 11 | 888 | 0.028571429 | 0.0044790407 | 2.673311 | AGUGUG,GAGUGU,GUGUGG,GUGUGU,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPLL | 7 | 748 | 0.019047619 | 0.0037736800 | 2.335567 | ACACAC,CACACA,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| CELF5 | 6 | 669 | 0.016666667 | 0.0033756550 | 2.303726 | GUGUGG,GUGUGU,GUGUUU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SSB | 4 | 505 | 0.011904762 | 0.0025493753 | 2.223323 | CUGUUU,GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPH3 | 22 | 2496 | 0.054761905 | 0.0125806127 | 2.121970 | AGGGAA,AUUGGG,GAAGGG,GAGGGG,GGAAUG,GGAGGG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGC,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CELF6 | 10 | 1196 | 0.026190476 | 0.0060308343 | 2.118613 | GUGGGG,GUGGUG,GUGUGG,UGUGAG,UGUGGG,UGUGGU,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| CELF4 | 6 | 776 | 0.016666667 | 0.0039147521 | 2.089973 | GUGUGG,GUGUGU,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPH2 | 27 | 3198 | 0.066666667 | 0.0161174929 | 2.048338 | AAGGGG,AGGGAA,AUUGGG,CUGGGG,GAAGGG,GAGGGG,GGAAUG,GGAGGG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,UGGGGG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 27 | 3406 | 0.066666667 | 0.0171654575 | 1.957457 | AAGGGG,AGGGAA,AUUGGG,CUGGGG,GAAGGG,GAGGGG,GGAAUG,GGAGGG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,UGGGGG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPF | 22 | 3360 | 0.054761905 | 0.0169336961 | 1.693276 | AAGGGG,AGGGAA,CUGGGG,GAAGGG,GAGGGG,GGAAUG,GGAGGG,GGGAAU,GGGAGG,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAU,GGGGAG,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| YBX1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | CCACAC,CCCUGC,CCUGCG,CUGCGG,GCCUGC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CUGGAC,GCGGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| HNRNPA2B1 | 9 | 2033 | 0.023809524 | 0.0102478839 | 1.216213 | AAGGGG,AGAAGC,AGGGGC,GAAGCC,GGAGCC,GGGGCC,UAGGGA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| MBNL1 | 15 | 3258 | 0.038095238 | 0.0164197904 | 1.214175 | CCUGCU,CUGCCU,CUGCGG,CUGCUU,GCUGCU,GCUUGC,GCUUUU,GGCUUU,GUGCUG,UGCUGU,UGCUUU,UUGCCU,UUGCUG | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUGU,GUGUUU,UGUGUG,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| SNRPA | 8 | 1947 | 0.021428571 | 0.0098145909 | 1.126536 | AUGCAC,CCUGCU,GUUUCC,UGCACA,UGCACC,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | ACACAC,CACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | GGAGGG,GGAGUA,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.