circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000102024:+:X:115610242:115610323
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000102024:+:X:115610242:115610323 | ENSG00000102024 | ENST00000481823 | + | X | 115610243 | 115610323 | 81 | AUCUUUAAAUGGAUGAGAUGGCUACCACUCAGAUUUCCAAAGAUGAGCUUGAUGAACUCAAAGAGGCCUUUGCAAAAGUUGAUCUUUAAAUGGAUGAGAUGGCUACCACUCAGAUUUCCAAAGAUGAGCUU | circ |
| ENSG00000102024:+:X:115610242:115610323 | ENSG00000102024 | ENST00000481823 | + | X | 115610243 | 115610323 | 22 | UGCAAAAGUUGAUCUUUAAAUG | bsj |
| ENSG00000102024:+:X:115610242:115610323 | ENSG00000102024 | ENST00000481823 | + | X | 115610043 | 115610252 | 210 | UUUUCUCAAUAAAUUUUGGUAAGUGAUGACUUCUCAUUUGUUUUCUUUAUUAAAGCUGUAUGUAGAGACAUGGUAACCUAUUCAGAUUGUUUUGAUAGUUUGAAAUUCUGUUUAAAUUAUCCCUAAGAUGAGAACUUAGCAAGAGUGCUUAUAUUUAUCACAAUUUUUUAAAGUCUGAAACGUUUUUGGUUUUUCUUUAGAUCUUUAAAU | ie_up |
| ENSG00000102024:+:X:115610242:115610323 | ENSG00000102024 | ENST00000481823 | + | X | 115610314 | 115610523 | 210 | GCAAAAGUUGGUGAGUAUUUUUUGUAGUAAAACUAUAGGGAAGCAAUAUUUAUUUGGUCUCAAAGUUAUUUCUCUAUAAAUAUCACAUGAAUGGUUAAUCAAAAACAUUAGGGCAAGAUAGUUAAAAUAGAUGGGAUAAAGUUUUGGAAGCUAUACUUAAAAUUGUUUUAUUUCUUUUUUUUUUUUUUGGUCGAGUGGUAGCUGAUAUUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
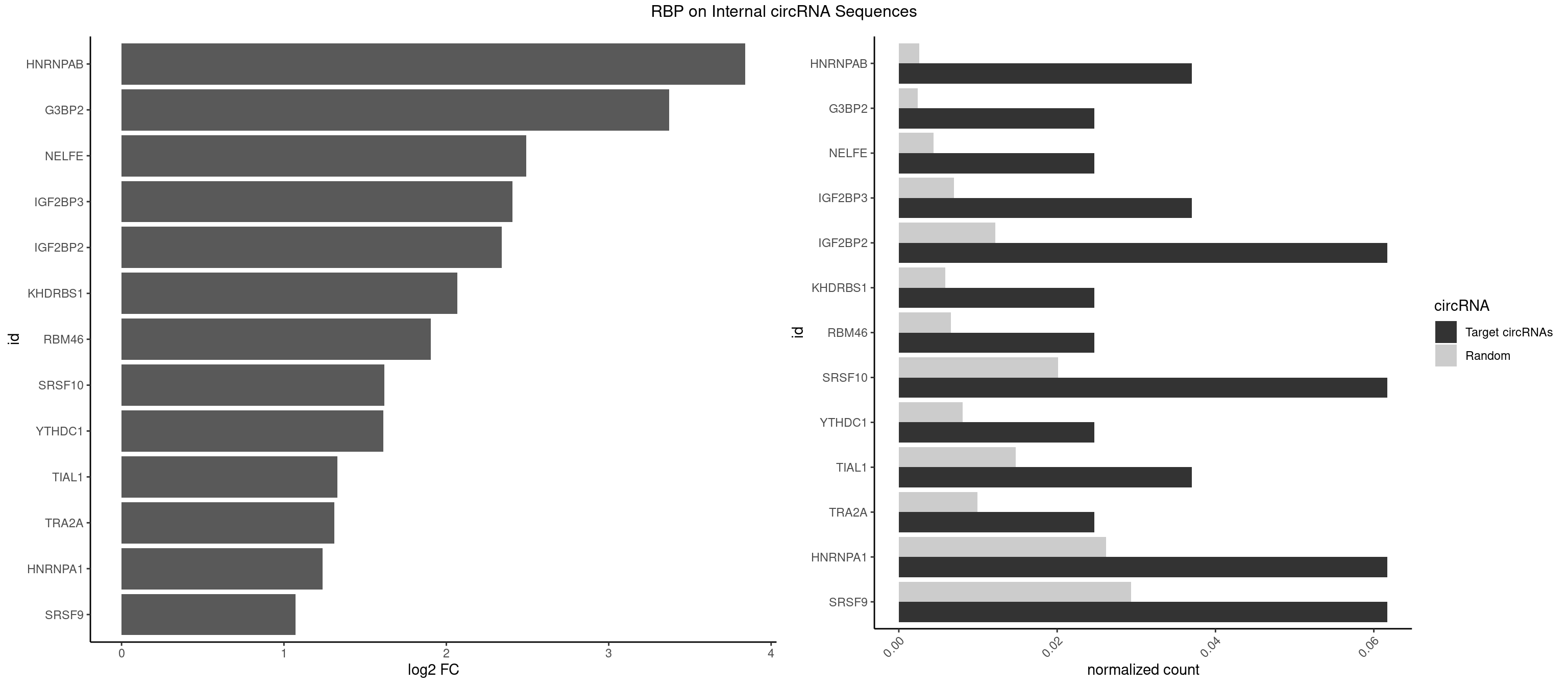
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPAB | 2 | 1782 | 0.03703704 | 0.002583931 | 3.841329 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| G3BP2 | 1 | 1644 | 0.02469136 | 0.002383941 | 3.372586 | GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| NELFE | 1 | 3028 | 0.02469136 | 0.004389639 | 2.491832 | UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| IGF2BP3 | 2 | 4815 | 0.03703704 | 0.006979366 | 2.407801 | AACUCA,CACUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| IGF2BP2 | 4 | 8408 | 0.06172840 | 0.012186356 | 2.340667 | AACUCA,CACUCA,CCACUC,GAACUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| KHDRBS1 | 1 | 4064 | 0.02469136 | 0.005891014 | 2.067418 | UUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBM46 | 1 | 4554 | 0.02469136 | 0.006601124 | 1.903223 | GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SRSF10 | 4 | 13860 | 0.06172840 | 0.020087416 | 1.619642 | AAAGAG,AAGAGG,CAAAGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| YTHDC1 | 1 | 5576 | 0.02469136 | 0.008082210 | 1.611184 | GGCUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| TIAL1 | 2 | 10182 | 0.03703704 | 0.014757244 | 1.327545 | UUAAAU,UUUAAA | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TRA2A | 1 | 6871 | 0.02469136 | 0.009958930 | 1.309944 | AAGAGG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| HNRNPA1 | 4 | 18070 | 0.06172840 | 0.026188565 | 1.236997 | AAAGAG,AAGAGG,CAAAGA | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| SRSF9 | 4 | 20254 | 0.06172840 | 0.029353626 | 1.072396 | AUGAAC,AUGAGA,AUGAGC,GAUGGC | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
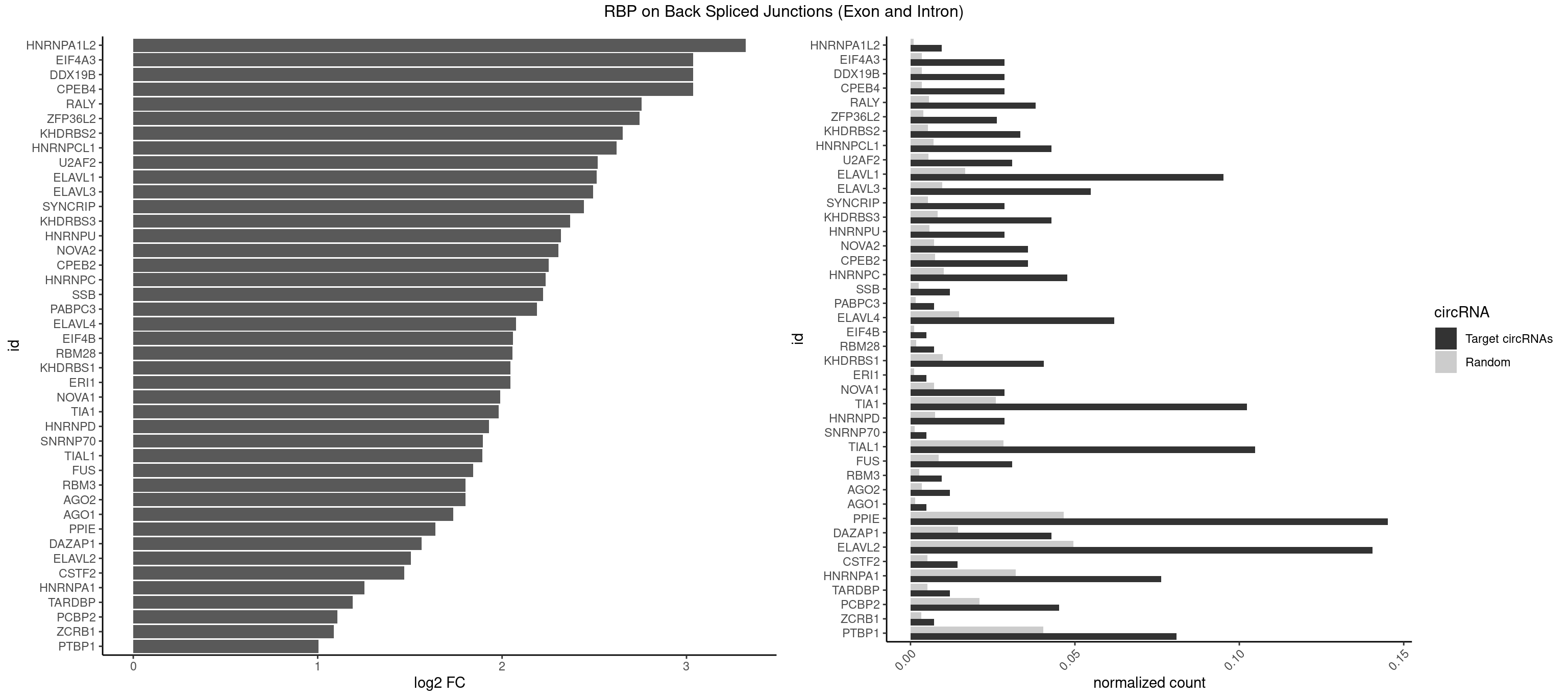
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 3 | 188 | 0.009523810 | 0.000952237 | 3.322146 | AUAGGG,UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| CPEB4 | 11 | 691 | 0.028571429 | 0.003486497 | 3.034723 | UUUUUU | UUUUUU |
| DDX19B | 11 | 691 | 0.028571429 | 0.003486497 | 3.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 11 | 691 | 0.028571429 | 0.003486497 | 3.034723 | UUUUUU | UUUUUU |
| RALY | 15 | 1117 | 0.038095238 | 0.005632809 | 2.757684 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ZFP36L2 | 10 | 774 | 0.026190476 | 0.003904676 | 2.745768 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| KHDRBS2 | 13 | 1051 | 0.033333333 | 0.005300282 | 2.652825 | AAUAAA,GAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPCL1 | 17 | 1381 | 0.042857143 | 0.006962918 | 2.621772 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| U2AF2 | 12 | 1071 | 0.030952381 | 0.005401048 | 2.518739 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| ELAVL1 | 39 | 3309 | 0.095238095 | 0.016676743 | 2.513701 | AUUUAU,UAGUUU,UAUUUA,UAUUUU,UGGUUU,UGUUUU,UUAUUU,UUGGUU,UUGUUU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ELAVL3 | 22 | 1928 | 0.054761905 | 0.009718863 | 2.494313 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| SYNCRIP | 11 | 1041 | 0.028571429 | 0.005249899 | 2.444212 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS3 | 17 | 1646 | 0.042857143 | 0.008298065 | 2.368689 | AAUAAA,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPU | 11 | 1136 | 0.028571429 | 0.005728537 | 2.318335 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| NOVA2 | 14 | 1434 | 0.035714286 | 0.007229948 | 2.304444 | AGACAU,GAGACA,UAGAUC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| CPEB2 | 14 | 1487 | 0.035714286 | 0.007496977 | 2.252120 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPC | 19 | 2006 | 0.047619048 | 0.010111850 | 2.235492 | AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SSB | 4 | 505 | 0.011904762 | 0.002549375 | 2.223323 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| PABPC3 | 2 | 310 | 0.007142857 | 0.001566909 | 2.188580 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ELAVL4 | 25 | 2916 | 0.061904762 | 0.014696695 | 2.074559 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| EIF4B | 1 | 226 | 0.004761905 | 0.001143692 | 2.057840 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM28 | 2 | 340 | 0.007142857 | 0.001718057 | 2.055723 | UGUAGA,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| KHDRBS1 | 16 | 1945 | 0.040476190 | 0.009804514 | 2.045555 | UAAAAC,UUAAAA,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ERI1 | 1 | 228 | 0.004761905 | 0.001153769 | 2.045185 | UUCAGA | UUCAGA,UUUCAG |
| NOVA1 | 11 | 1428 | 0.028571429 | 0.007199718 | 1.988561 | UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| TIA1 | 42 | 5140 | 0.102380952 | 0.025901854 | 1.982820 | AUUUUG,AUUUUU,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPD | 11 | 1488 | 0.028571429 | 0.007502015 | 1.929223 | AUUUAU,UAUUUA,UUAGGG,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.001279726 | 1.895704 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| TIAL1 | 43 | 5597 | 0.104761905 | 0.028204353 | 1.893124 | AAAUUU,AAUUUU,AGUUUU,AUUUUG,AUUUUU,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| FUS | 12 | 1711 | 0.030952381 | 0.008625554 | 1.843361 | UGGUGA,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| RBM3 | 3 | 541 | 0.009523810 | 0.002730754 | 1.802240 | AAAACU,AAACUA,GAAACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| AGO2 | 4 | 677 | 0.011904762 | 0.003415961 | 1.801175 | AGUGCU,GUGCUU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| AGO1 | 1 | 283 | 0.004761905 | 0.001430875 | 1.734641 | GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| PPIE | 60 | 9262 | 0.145238095 | 0.046669690 | 1.637862 | AAAAUA,AAAAUU,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAUU,AAUUAU,AAUUUU,AUAAAU,AUAUUA,AUAUUU,AUUAAA,AUUUAU,AUUUUU,UAAAAU,UAAAUA,UAAAUU,UAUAAA,UAUAUU,UAUUAA,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| DAZAP1 | 17 | 2878 | 0.042857143 | 0.014505240 | 1.562962 | AGUAAA,AGUUAA,AGUUUG,UAGUAA,UAGUUA,UAGUUU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ELAVL2 | 58 | 9826 | 0.140476190 | 0.049511286 | 1.504496 | AAUUUU,AUACUU,AUAUUU,AUUUAU,AUUUUG,AUUUUU,CUUAUA,CUUUAA,CUUUUU,GUUUUA,GUUUUC,GUUUUU,UAGUUA,UAUACU,UAUAUU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUUU,UUAUAU,UUAUUU,UUCUUU,UUUAUU,UUUCUU,UUUGAU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.005154172 | 1.470761 | GUUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| HNRNPA1 | 31 | 6340 | 0.076190476 | 0.031947803 | 1.253894 | AGGGCA,AGUGGU,AUAGGG,AUUUAU,GAGUGG,GUAAGU,UAGAGA,UAGGGA,UAGGGC,UAGUUA,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUU,UUUUUU | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.005214631 | 1.190902 | GAAUGG,GUUUUG,UGAAUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| PCBP2 | 18 | 4164 | 0.045238095 | 0.020984482 | 1.108215 | AAAUUA,AAAUUC,ACAUUA,AUUAAA,CCCUAA,CUUAAA,UUAGGG,UUUUUU | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.003360540 | 1.087808 | ACUUAA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PTBP1 | 33 | 8003 | 0.080952381 | 0.040326481 | 1.005346 | AGCUGU,AUUUUU,CUUCUC,CUUUUU,UAGCUG,UAUUUU,UCUUUU,UUAUUU,UUCUCU,UUCUUU,UUUCUU,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.