circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000110911:-:12:50990794:51010766
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000110911:-:12:50990794:51010766 | ENSG00000110911 | ENST00000394904 | - | 12 | 50990795 | 51010766 | 1613 | AAUCAUAUUCUAAGAACUCAGCCACUCAGGUAUCCACCAUGGUGCUGGGUCCUGAACAGAAGAUGUCAGAUGACAGUGUUUCUGGAGAUCAUGGGGAGUCUGCCAGUCUUGGUAACAUCAACCCUGCCUAUAGUAAUCCCUCUCUUUCACAGUCCCCUGGGGACUCAGAGGAGUACUUCGCCACUUACUUUAAUGAGAAGAUCUCCAUUCCUGAGGAGGAGUACUCUUGUUUUAGCUUUCGUAAACUCUGGGCUUUCACCGGACCAGGUUUUCUUAUGAGCAUUGCCUACCUGGAUCCAGGAAAUAUUGAAUCCGAUUUGCAGUCUGGAGCAGUGGCUGGAUUUAAGUUGCUCUGGAUCCUUCUGUUGGCCACCCUUGUGGGGCUGCUGCUCCAGCGGCUUGCAGCUAGACUGGGAGUGGUUACUGGGCUGCAUCUUGCUGAAGUAUGUCACCGUCAGUAUCCCAAGGUCCCACGAGUCAUCCUGUGGCUGAUGGUGGAGUUGGCUAUCAUCGGCUCAGACAUGCAAGAAGUCAUUGGCUCAGCCAUUGCUAUCAAUCUUCUGUCUGUAGGAAGAAUUCCUCUGUGGGGUGGCGUUCUCAUCACCAUUGCAGAUACUUUUGUAUUUCUCUUCUUGGACAAAUAUGGCUUGCGGAAGCUAGAAGCAUUUUUUGGCUUUCUCAUCACUAUUAUGGCCCUCACAUUUGGAUAUGAGUAUGUUACAGUGAAACCCAGCCAGAGCCAGGUACUCAAGGGCAUGUUCGUACCAUCCUGUUCAGGCUGUCGCACUCCACAGAUUGAACAGGCUGUGGGCAUCGUGGGAGCUGUCAUCAUGCCACACAACAUGUACCUGCAUUCUGCCUUAGUCAAGUCUAGACAGGUAAACCGGAACAAUAAGCAGGAAGUUCGAGAAGCCAAUAAGUACUUUUUCAUUGAAUCCUGCAUUGCACUCUUUGUUUCCUUCAUCAUCAAUGUCUUUGUUGUCUCAGUCUUUGCUGAAGCAUUUUUUGGGAAAACCAACGAGCAGGUGGUUGAAGUCUGUACAAAUACCAGCAGUCCUCAUGCUGGCCUCUUUCCUAAAGAUAACUCGACACUGGCUGUGGACAUCUACAAAGGGGGUGUUGUGCUGGGAUGUUACUUUGGGCCUGCUGCACUCUACAUUUGGGCAGUGGGGAUCCUGGCUGCAGGACAGAGCUCCACCAUGACAGGAACCUAUUCUGGCCAGUUUGUCAUGGAGGGAUUCCUGAACCUAAAGUGGUCACGCUUUGCCCGAGUGGUUCUGACUCGCUCUAUUGCCAUCAUCCCCACUCUGCUUGUUGCUGUCUUCCAAGAUGUAGAGCAUCUAACAGGGAUGAAUGACUUUCUGAAUGUUCUACAGAGCUUACAGCUUCCCUUUGCUCUCAUACCCAUCCUCACAUUUACGAGCUUGCGGCCAGUAAUGAGUGACUUUGCCAAUGGACUAGGCUGGCGGAUUGCAGGAGGAAUCUUGGUCCUUAUCAUCUGUUCCAUCAAUAUGUACUUUGUAGUGGUUUAUGUCCGGGACCUAGGGCAUGUGGCAUUAUAUGUGGUGGCUGCUGUGGUCAGCGUGGCUUAUCUGGGCUUUGUGUUCUACUUGAAUCAUAUUCUAAGAACUCAGCCACUCAGGUAUCCACCAUGGUGCUGGGU | circ |
| ENSG00000110911:-:12:50990794:51010766 | ENSG00000110911 | ENST00000394904 | - | 12 | 50990795 | 51010766 | 22 | UGUUCUACUUGAAUCAUAUUCU | bsj |
| ENSG00000110911:-:12:50990794:51010766 | ENSG00000110911 | ENST00000394904 | - | 12 | 51010757 | 51010966 | 210 | UUGACUAAGACUUUUCUUUUUUUGUUAAAUAUCUGUCAGUCAUAUUCUGAAAACUGUAAUUAUUUUCAAGGUUACCUAAUUCAGUAGAAAAAUUACUCUUUUCAUAGUAAGGAUUCAGUCAUACAGAGUAACAAACUGCUGGUUCUGAACUUGUUAGUUUGUUCUUCCUAAUUCUUCUCCCUCACUUUUCUCCUUUCCAGAAUCAUAUUC | ie_up |
| ENSG00000110911:-:12:50990794:51010766 | ENSG00000110911 | ENST00000394904 | - | 12 | 50990595 | 50990804 | 210 | GUUCUACUUGGUAAGUCUAGCCUGGAAGAGCAGGGGCAUAGGUAUCAGGGAUGGUUGAGGUGGGGUUCACUGACAUUCAGAAAUGAACAGACCCAGGCUGGGCACAGUGGCUCAUGCCUAUAAUCCCAGUACUUUGGGAGGCUGAGGCGGGAGGAUUGCUUGAAUCCAGGAGUUAGAGACCAAUCUGGGCAACAUAGUAAGACUCCAUCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
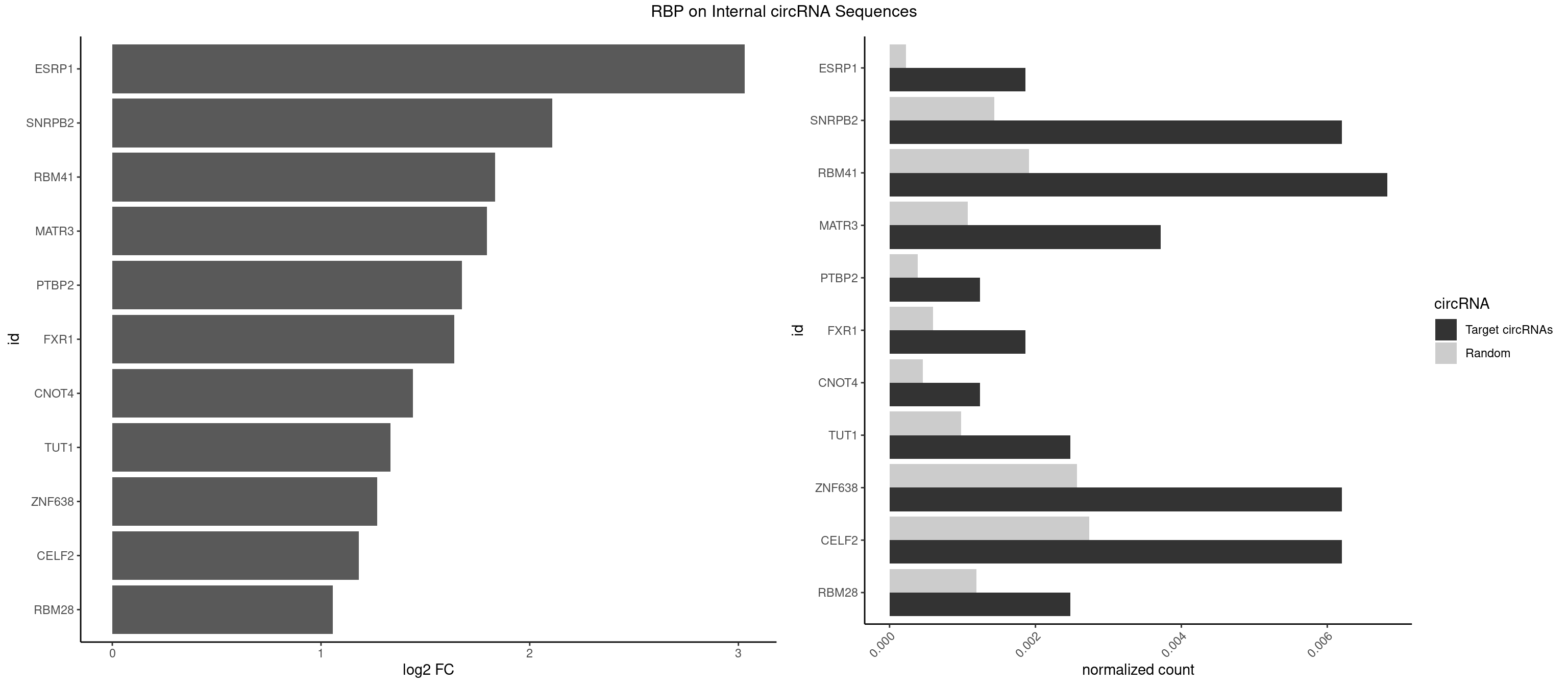
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 2 | 156 | 0.001859888 | 0.0002275250 | 3.031119 | AGGGAU | AGGGAU |
| SNRPB2 | 9 | 991 | 0.006199628 | 0.0014376103 | 2.108509 | AUUGCA,UAUUGC,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM41 | 10 | 1318 | 0.006819591 | 0.0019115000 | 1.834980 | AUACUU,UACAUU,UACUUG,UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| MATR3 | 5 | 739 | 0.003719777 | 0.0010724109 | 1.794358 | AAUCUU,AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| PTBP2 | 1 | 267 | 0.001239926 | 0.0003883867 | 1.674688 | CUCUCU | CUCUCU |
| FXR1 | 2 | 411 | 0.001859888 | 0.0005970720 | 1.639239 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| CNOT4 | 1 | 314 | 0.001239926 | 0.0004564992 | 1.441569 | GACAGA | GACAGA |
| TUT1 | 3 | 678 | 0.002479851 | 0.0009840095 | 1.333509 | AAAUAC,AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ZNF638 | 9 | 1773 | 0.006199628 | 0.0025708878 | 1.269915 | CGUUCU,GGUUCU,GUUCGU,UGUUCG,UGUUCU,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF2 | 9 | 1886 | 0.006199628 | 0.0027346479 | 1.180827 | GUAUGU,GUCUGU,UAUGUG,UAUGUU,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM28 | 3 | 822 | 0.002479851 | 0.0011926949 | 1.056029 | UGUAGA,UGUAGG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
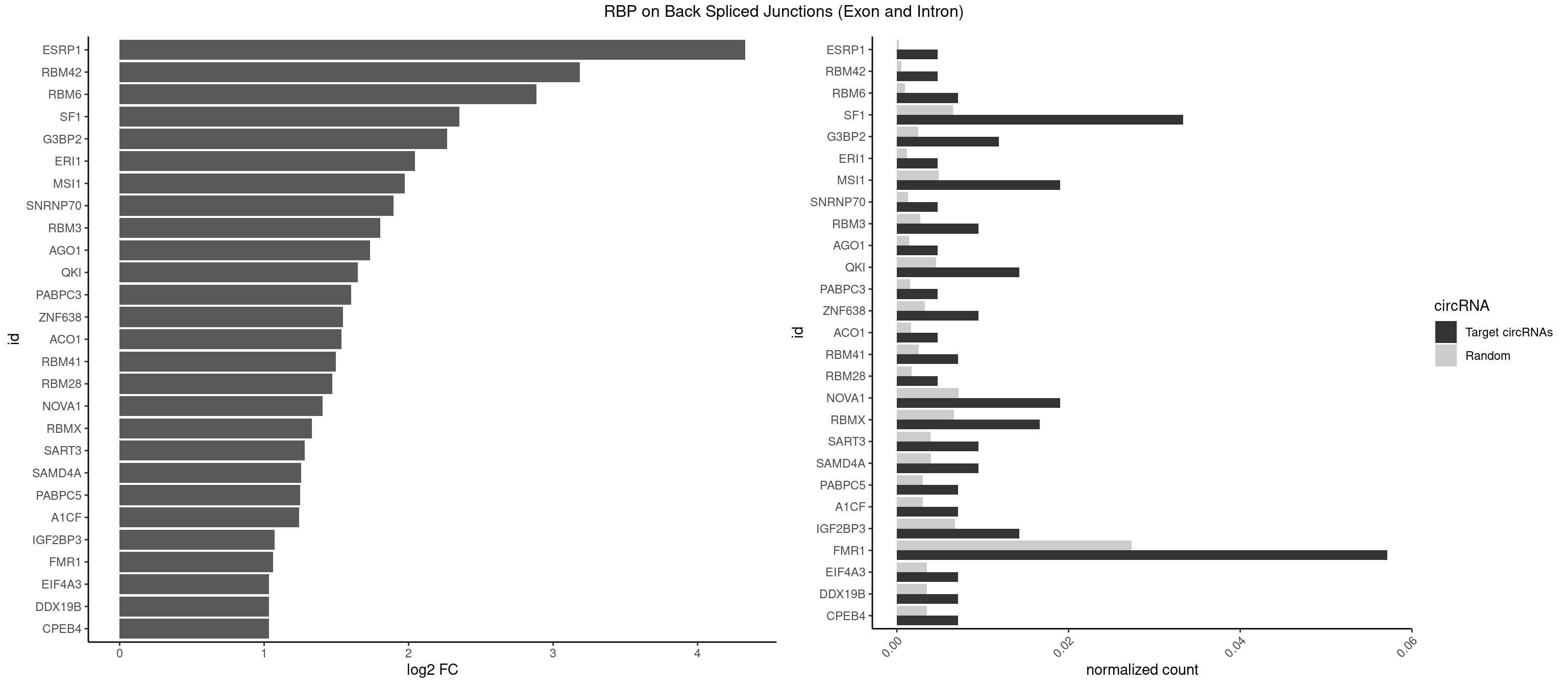
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 46 | 0.004761905 | 0.0002367997 | 4.329800 | AGGGAU | AGGGAU |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| SF1 | 13 | 1294 | 0.033333333 | 0.0065245869 | 2.353007 | ACAGAC,ACUAAG,ACUGAC,AGUAAC,AGUAAG,CACUGA,CAGUCA,GACUAA,UAACAA,UAGUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| G3BP2 | 4 | 490 | 0.011904762 | 0.0024738009 | 2.266737 | AGGAUU,GGAUGG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUCAGA | UUCAGA,UUUCAG |
| MSI1 | 7 | 961 | 0.019047619 | 0.0048468360 | 1.974496 | AGGUGG,AGUAAG,AGUUAG,UAGGUA,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | AAUCAU,AUCAUA,UCAUAU,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUCU,GUUCUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACUUG,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| NOVA1 | 7 | 1428 | 0.019047619 | 0.0071997179 | 1.403598 | CAGUCA,CAUUCA,UCAGUC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| RBMX | 6 | 1316 | 0.016666667 | 0.0066354293 | 1.328704 | AGUAAC,AUCCCA,GUAACA,UAACAA,UAAGAC | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CUGGAA,GCGGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | UAAUUA,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAUUCA,ACAAAC,ACAUUC,CAUACA,CAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| FMR1 | 23 | 5430 | 0.057142857 | 0.0273629585 | 1.062349 | AAGGAU,ACUAAG,AGGAGU,AGGAUU,AGUGGC,CAUAGG,CUGAGG,CUGGGC,GACUAA,GAGGAU,GCAUAG,GCUGAG,GCUGGG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GUGGCU,UAAGGA,UUUUUU | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.