circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000099203:-:19:10834933:10836430
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000099203:-:19:10834933:10836430 | ENSG00000099203 | MSTRG.16429.1 | - | 19 | 10834934 | 10836430 | 704 | CGGCCGAGAAGAGAGCCGAAGCCUUUGAGAAAAAUAUUCCCAGACGGUUGAGUGUUUCUGGGCACAUCAGGCAUUCAUUCGGGCCGCCUGCCCUCUAGUGGGGGGGUCCUUCACUCCUGGCACCUCUGGUUACGGACAACAUACAGAAGCCACUGCCAAUGAGCUCCGCCGAGUAGCACCGGGGCAGGGCUAGCGCUUAAAGGAGCCGCGACCCCUUUGCAGACCAGAGGGUGACCCGGAUGAUGGCGGCCGGCGCGGCCCUAGCCCUGGCCUUGUGGCUACUAAUGCCACCAGUGGAGGUGGGAGGGGCGGGGCCCCCGCCAAUCCAGGACGGUGAGUUCACGUUCCUGUUGCCGGCGGGGAGGAAGCAGUGUUUCUACCAGUCCGCGCCGGCCAACGCAAGCCUCGAGACCGAAUACCAGGUGAUCGGAGGUGCUGGACUGGACGUGGACUUCACGCUGGAGAGCCCUCAGGGCGUGCUGUUGGUCAGCGAGUCCCGCAAGGCUGAUGGGGUACACACGGUGGAGCCAACGGAGGCCGGGGACUACAAGCUGUGCUUUGACAACUCCUUCAGCACCAUCUCCGAGAAGCUGGUGUUCUUUGAACUGAUCUUUGACAGCCUCCAGGAUGACGAGGAGGUCGAAGGAUGGGCAGAGGCUGUGGAGCCCGAGGAGAUGCUGGAUGUUAAAAUGGAGGACAUCAAGCGGCCGAGAAGAGAGCCGAAGCCUUUGAGAAAAAUAUUCCCAGACGGUUG | circ |
| ENSG00000099203:-:19:10834933:10836430 | ENSG00000099203 | MSTRG.16429.1 | - | 19 | 10834934 | 10836430 | 22 | AGGACAUCAAGCGGCCGAGAAG | bsj |
| ENSG00000099203:-:19:10834933:10836430 | ENSG00000099203 | MSTRG.16429.1 | - | 19 | 10836421 | 10836630 | 210 | CAGGGCCACGCCUAGACGCUAUUUUUGGGUCCACUCCACCACGUCGGUCCCGCCAAAGGUUGUCACGCGCGGCGCACUCUGGGAAAACCACGUGGAGGUGACUAAAAAGGACCCUGCCGCCUGUAACUUUAGUUGUCUUAGCUACUGCCUACAAAAGAUGCGCCAUCAUGGCACUUAUUUUUGUAAUUUUCCUCCACCAGCGGCCGAGAA | ie_up |
| ENSG00000099203:-:19:10834933:10836430 | ENSG00000099203 | MSTRG.16429.1 | - | 19 | 10834734 | 10834943 | 210 | GGACAUCAAGGUGUGUCCAGGCUGCGCUGGGGCCACUCCUUCCAGAUCUCUAGGAUUCACCUGGGGUGCCCCCCUUGGGCUGACCCAGCCUCCCCUCCGUAGUUGUGCCCUCCAACUGAAUUAUAGUAAGAGUAGCAGCUCUUAAAAUAAGUAGUUCAUUUCAGGCCGGGCACGGUGGCUCACGCCUGUAAUCCCAGCACUUUGGGAGGC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
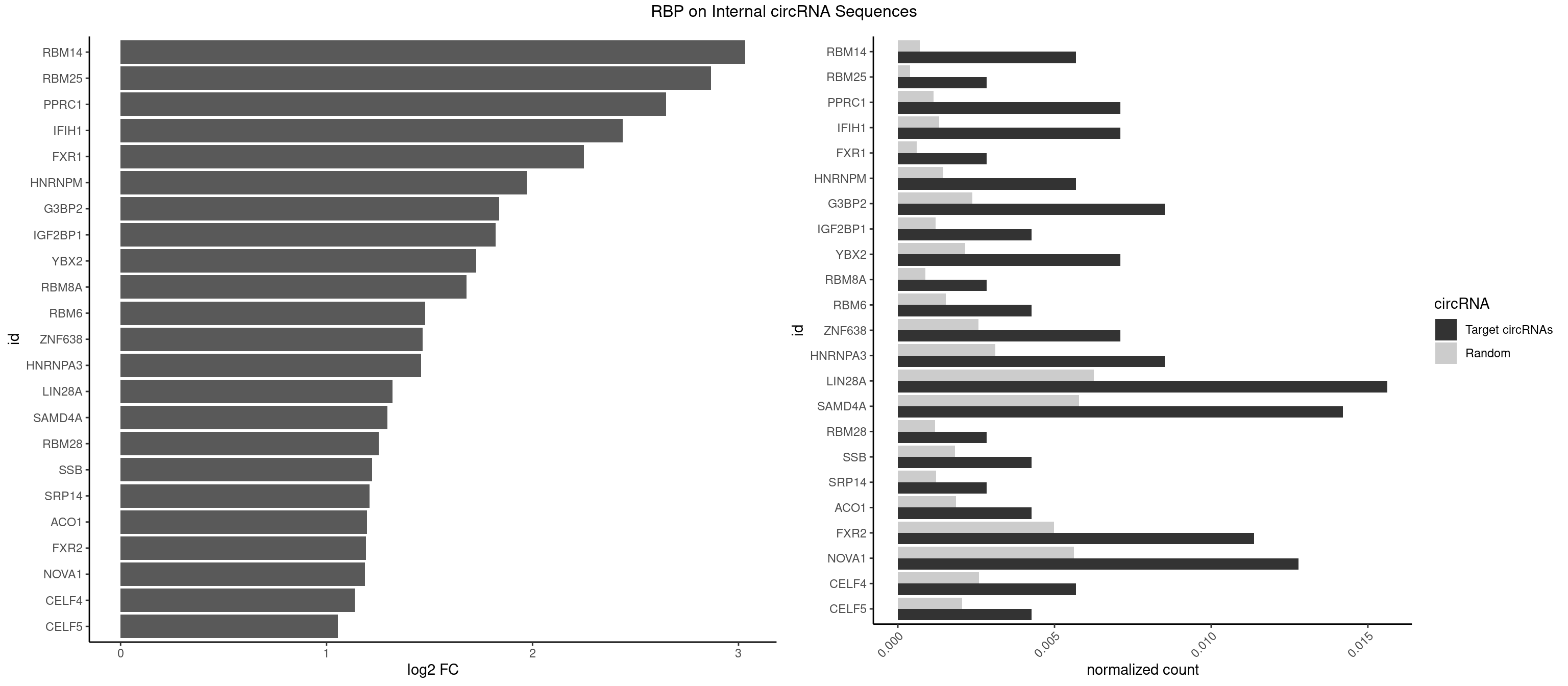
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM14 | 3 | 478 | 0.005681818 | 0.0006941687 | 3.032994 | CGCGCC,CGCGGC,GCGCGG | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBM25 | 1 | 268 | 0.002840909 | 0.0003898359 | 2.865414 | UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| PPRC1 | 4 | 780 | 0.007102273 | 0.0011318283 | 2.649626 | CCGCGC,CGCGCC,CGGCGC,GGCGCG | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| IFIH1 | 4 | 904 | 0.007102273 | 0.0013115296 | 2.437030 | GCCGCG,GGCCCU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| FXR1 | 1 | 411 | 0.002840909 | 0.0005970720 | 2.250376 | AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| HNRNPM | 3 | 999 | 0.005681818 | 0.0014492040 | 1.971092 | GAAGGA,GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 5 | 1644 | 0.008522727 | 0.0023839405 | 1.837967 | AGGAUG,GGAUGA,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| IGF2BP1 | 2 | 831 | 0.004261364 | 0.0012057377 | 1.821399 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| YBX2 | 4 | 1480 | 0.007102273 | 0.0021462711 | 1.726448 | ACAACA,ACAACU,ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM8A | 1 | 611 | 0.002840909 | 0.0008869128 | 1.679488 | CGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| RBM6 | 2 | 1054 | 0.004261364 | 0.0015289102 | 1.478812 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ZNF638 | 4 | 1773 | 0.007102273 | 0.0025708878 | 1.466014 | GUUCUU,GUUGGU,UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPA3 | 5 | 2140 | 0.008522727 | 0.0031027457 | 1.457770 | AAGGAG,AGGAGC,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| LIN28A | 10 | 4315 | 0.015625000 | 0.0062547643 | 1.320829 | AGGAGA,CGGAGG,GGAGAU,GGAGGA,GGAGGG,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SAMD4A | 9 | 3992 | 0.014204545 | 0.0057866714 | 1.295547 | CGGGCC,CUGGAC,CUGGCA,CUGGCC,GCUGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| RBM28 | 1 | 822 | 0.002840909 | 0.0011926949 | 1.252128 | GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SSB | 2 | 1260 | 0.004261364 | 0.0018274462 | 1.221486 | GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SRP14 | 1 | 847 | 0.002840909 | 0.0012289250 | 1.208956 | CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ACO1 | 2 | 1283 | 0.004261364 | 0.0018607779 | 1.195409 | CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| FXR2 | 7 | 3434 | 0.011363636 | 0.0049780156 | 1.190782 | AGACGG,GACGAG,GGACAA,GGACGG,UGACAA,UGACAG,UGACGA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| NOVA1 | 8 | 3872 | 0.012784091 | 0.0056127669 | 1.187566 | AGCACC,AUUCAU,CAUUCA,GGGGGG,UCAUUC,UUCAUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| CELF4 | 3 | 1782 | 0.005681818 | 0.0025839306 | 1.136785 | GGUGUU,GUGUUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF5 | 2 | 1415 | 0.004261364 | 0.0020520728 | 1.054233 | GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
RBP on BSJ (Exon Only)
Plot
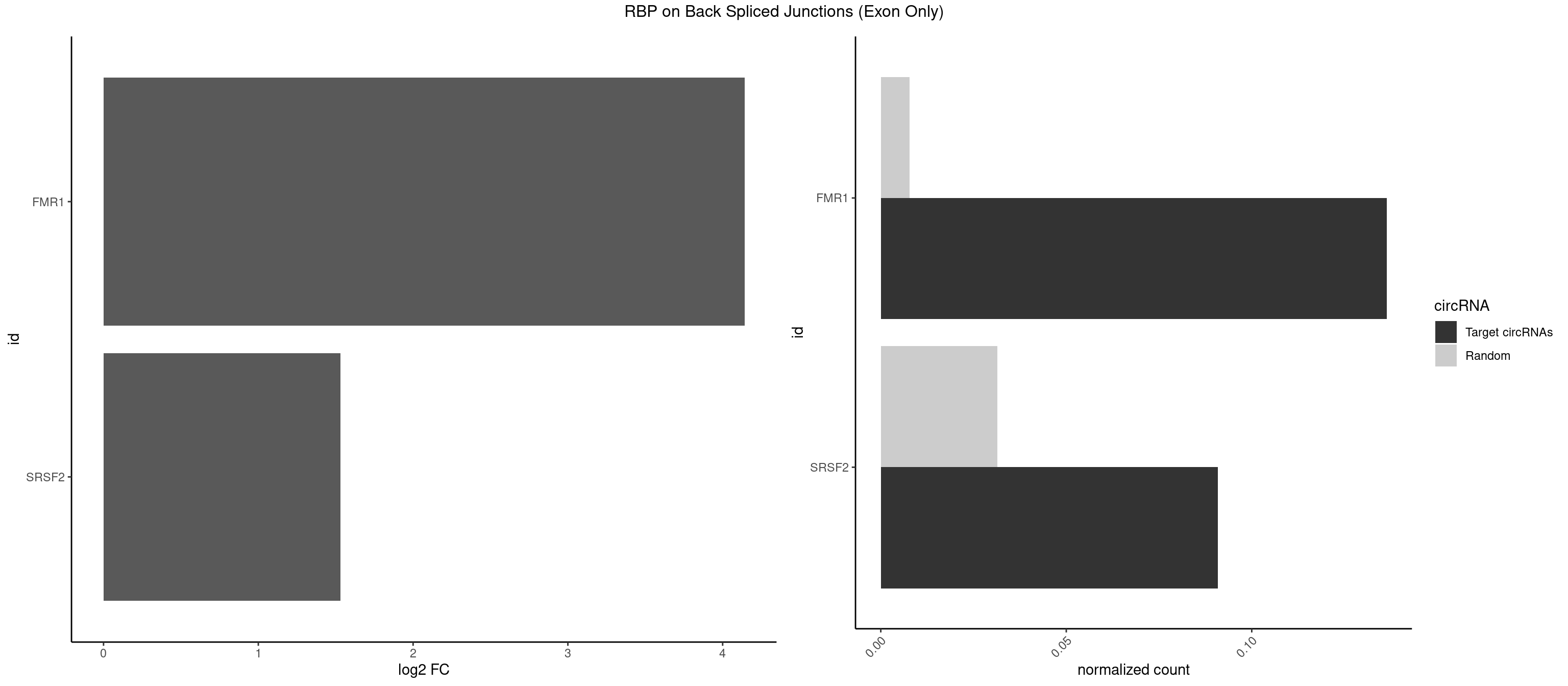
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FMR1 | 2 | 117 | 0.13636364 | 0.007728583 | 4.141111 | AAGCGG,AGCGGC | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF2 | 1 | 479 | 0.09090909 | 0.031438302 | 1.531901 | AAGCGG | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
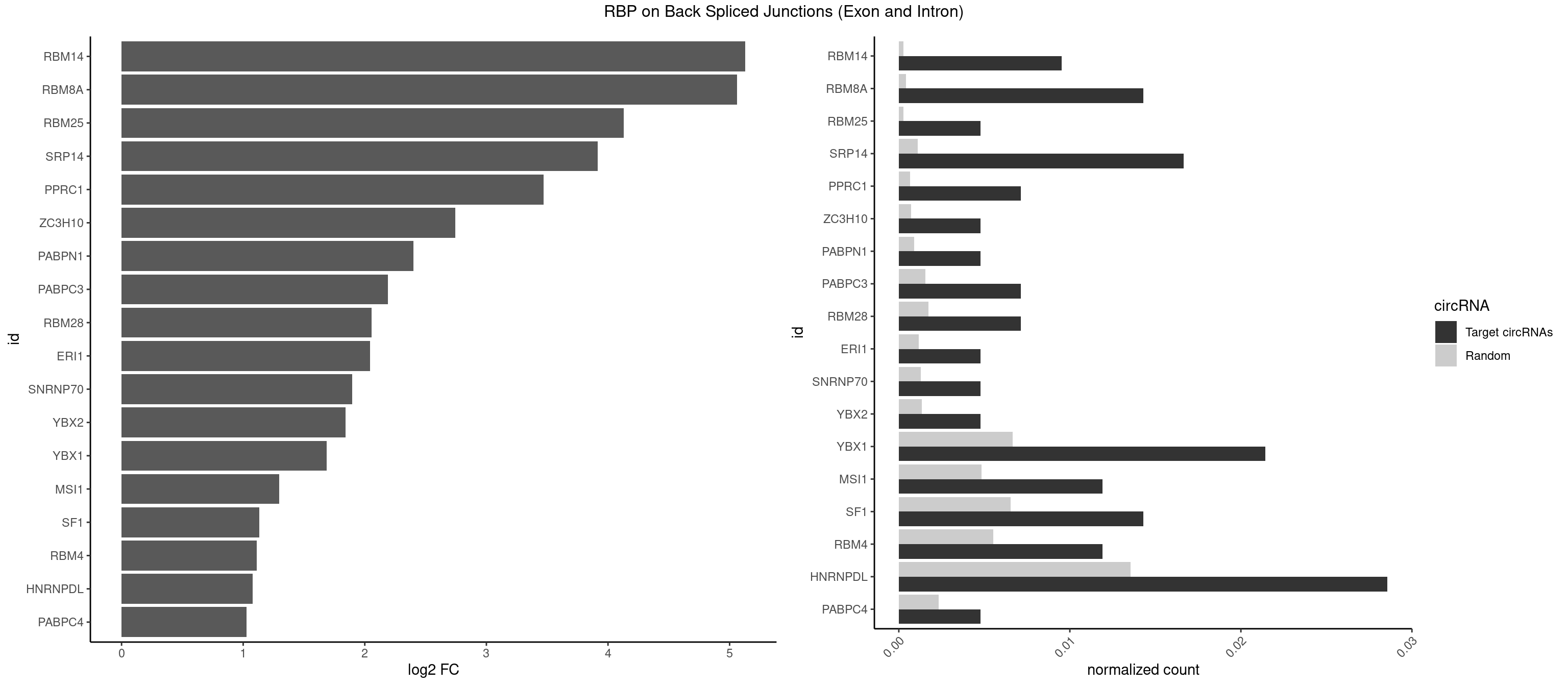
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM14 | 3 | 53 | 0.009523810 | 0.0002720677 | 5.129501 | CGCGCG,CGCGGC,GCGCGG | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBM8A | 5 | 84 | 0.014285714 | 0.0004282547 | 5.059960 | ACGCGC,AUGCGC,CGCGCG,UGCGCC,UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| RBM25 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| SRP14 | 6 | 218 | 0.016666667 | 0.0011033857 | 3.916956 | CCUGUA,CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| PPRC1 | 2 | 127 | 0.007142857 | 0.0006449012 | 3.469351 | CGCGCG,CGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | CCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGU,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| YBX1 | 8 | 1321 | 0.021428571 | 0.0066606207 | 1.685807 | AACCAC,ACAUCA,CACCAC,CAUCAU,CCACCA,CCAGCA,CCCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGUAAG,UAGUAA,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACUUAU,AGUAAG,GACUAA,GCUGAC,UAGUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUUCC,CGCGCG,GCGCGG,UCCUUC | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| HNRNPDL | 11 | 2689 | 0.028571429 | 0.0135530028 | 1.075961 | ACCACG,ACUUUA,AGUAGC,AUGCGC,CACCAG,CCACGC,CUAGGA,CUUUAG,GAGUAG,UAAGUA | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.