circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000114416:+:3:180968050:180970358
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000114416:+:3:180968050:180970358 | ENSG00000114416 | ENST00000305586 | + | 3 | 180968051 | 180970358 | 405 | GUACAAAUUCUGAGCUGUCUAACCCCUCUGAAACGGAAUCUGAGCGUAAAGACGAGCUGAGUGAUUGGUCAUUGGCAGGAGAAGAUGAUCGAGACAGCCGACAUCAGCGUGACAGCAGGAGACGCCCAGGAGGAAGAGGCAGAAGUGUUUCAGGGGGUCGAGGUCGUGGUGGACCACGUGGUGGCAAAUCCUCCAUCAGUUCUGUGCUCAAAGAUCCAGACAGCAAUCCAUACAGCUUACUUGAUAAUACAGAAUCAGAUCAGACUGCAGACACUGAUGCCAGCGAAUCUCAUCACAGUACUAACCGUCGUAGGCGGUCUCGUAGACGAAGGACUGAUGAAGAUGCUGUUCUGAUGGAUGGAAUGACUGAAUCUGAUACAGCUUCAGUUAAUGAAAAUGGGCUAGGUACAAAUUCUGAGCUGUCUAACCCCUCUGAAACGGAAUCUGAGCGUAAA | circ |
| ENSG00000114416:+:3:180968050:180970358 | ENSG00000114416 | ENST00000305586 | + | 3 | 180968051 | 180970358 | 22 | AAAUGGGCUAGGUACAAAUUCU | bsj |
| ENSG00000114416:+:3:180968050:180970358 | ENSG00000114416 | ENST00000305586 | + | 3 | 180967851 | 180968060 | 210 | AUACUAUACUAGUAUAUUAAUAGUAUAAUAAUUGAUUAUUCCUUAGUACGUUUCAGAUUGAUUUUAACUUUCAUGGAUCUUUCUUUGAAGCAGCCAAACAGUAUUGCUUAUUUAAACAAUAAUUUGUAGGACAUAAUUUUGAACAUUUUAAAAGGCUUUUUAUAGAAAUCUAAGUGGUUUAUGUUCUUUUCUUUUCCAAGGUACAAAUUC | ie_up |
| ENSG00000114416:+:3:180968050:180970358 | ENSG00000114416 | ENST00000305586 | + | 3 | 180970349 | 180970558 | 210 | AAUGGGCUAGGUAUGUAAGCACUUAGGGAAGAGAAAUAUAUAUAUAUAUAUAUAUAUAUAUAUAUAUAUAUAUAAUUGUAAACUAUUGUUAGUAUAUGAAGCUUUUUUCAAAGAAAUAUGCAAUUCACAAAUUGACUUUUUCUUAGUGUUUACUUUGAACAAGGUAGAUAGCUUUUUGUCUAUCUUAAAUGUUUAGUUCUCAGAUUUGUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
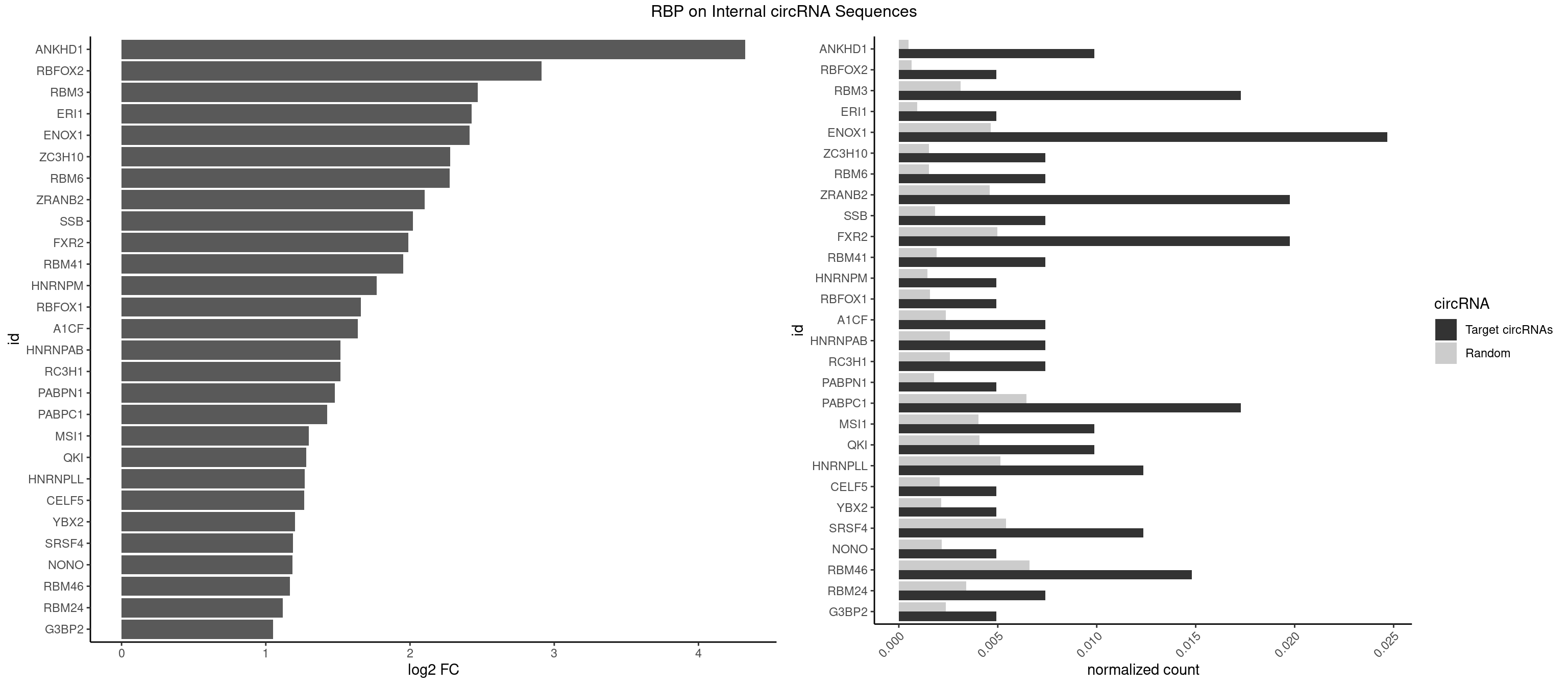
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 3 | 339 | 0.009876543 | 0.0004927293 | 4.325139 | AGACGA,GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBFOX2 | 1 | 452 | 0.004938272 | 0.0006564894 | 2.911163 | UGACUG | UGACUG,UGCAUG |
| RBM3 | 6 | 2152 | 0.017283951 | 0.0031201361 | 2.469752 | AAGACG,AGACGA,GAAACG,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ERI1 | 1 | 632 | 0.004938272 | 0.0009173461 | 2.428468 | UUUCAG | UUCAGA,UUUCAG |
| ENOX1 | 9 | 3195 | 0.024691358 | 0.0046316558 | 2.414406 | AAUACA,AGACAG,AUACAG,CAGACA,CAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ZC3H10 | 2 | 1053 | 0.007407407 | 0.0015274610 | 2.277833 | CAGCGA,CCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBM6 | 2 | 1054 | 0.007407407 | 0.0015289102 | 2.276465 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ZRANB2 | 7 | 3173 | 0.019753086 | 0.0045997733 | 2.102443 | AGGUAC,CGGUCU,GUAAAG,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| SSB | 2 | 1260 | 0.007407407 | 0.0018274462 | 2.019140 | GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| FXR2 | 7 | 3434 | 0.019753086 | 0.0049780156 | 1.988435 | AGACAG,AGACGA,GACGAA,GACGAG,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| RBM41 | 2 | 1318 | 0.007407407 | 0.0019115000 | 1.954263 | UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPM | 1 | 999 | 0.004938272 | 0.0014492040 | 1.768746 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBFOX1 | 1 | 1077 | 0.004938272 | 0.0015622419 | 1.660388 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| A1CF | 2 | 1642 | 0.007407407 | 0.0023810421 | 1.637376 | AUCAGU,GAUCAG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPAB | 2 | 1782 | 0.007407407 | 0.0025839306 | 1.519401 | AAAGAC,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RC3H1 | 2 | 1786 | 0.007407407 | 0.0025897275 | 1.516168 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PABPN1 | 1 | 1222 | 0.004938272 | 0.0017723764 | 1.478321 | AGAAGA | AAAAGA,AGAAGA |
| PABPC1 | 6 | 4443 | 0.017283951 | 0.0064402624 | 1.424242 | ACAAAU,ACUAAC,CAAAUC,CGAAUC,CUAACC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| MSI1 | 3 | 2770 | 0.009876543 | 0.0040157442 | 1.298339 | AGGAAG,AGGAGG,UAGGUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| QKI | 3 | 2805 | 0.009876543 | 0.0040664663 | 1.280231 | ACUAAC,CUAACC | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPLL | 4 | 3534 | 0.012345679 | 0.0051229360 | 1.268963 | ACUGCA,AGACGA,CAUACA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| CELF5 | 1 | 1415 | 0.004938272 | 0.0020520728 | 1.266924 | GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| YBX2 | 1 | 1480 | 0.004938272 | 0.0021462711 | 1.202174 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SRSF4 | 4 | 3740 | 0.012345679 | 0.0054214720 | 1.187250 | AGAAGA,AGGAAG,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| NONO | 1 | 1498 | 0.004938272 | 0.0021723567 | 1.184745 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM46 | 5 | 4554 | 0.014814815 | 0.0066011240 | 1.166257 | AAUGAA,AUGAAA,AUGAAG,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM24 | 2 | 2357 | 0.007407407 | 0.0034172229 | 1.116144 | GAGUGA,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| G3BP2 | 1 | 1644 | 0.004938272 | 0.0023839405 | 1.050658 | GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
RBP on BSJ (Exon Only)
Plot
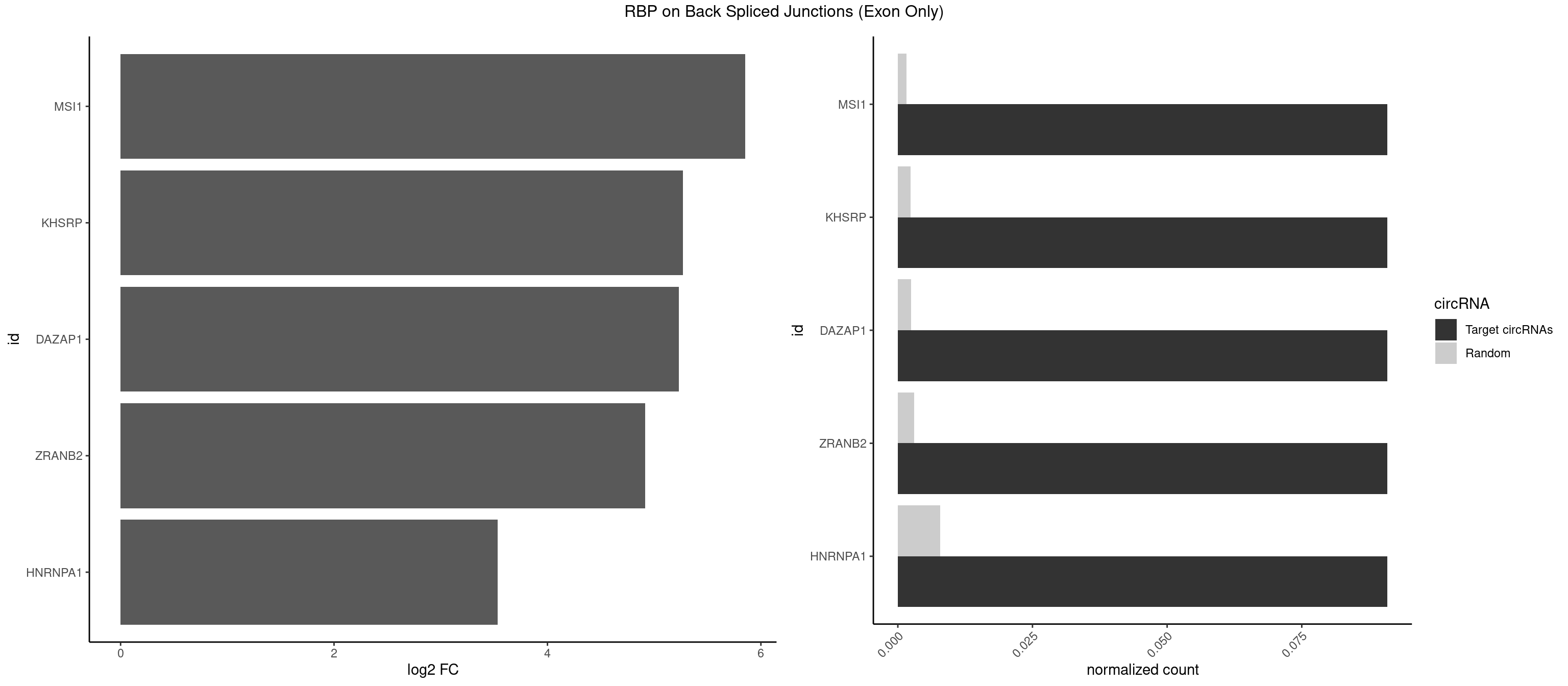
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MSI1 | 1 | 23 | 0.09090909 | 0.001571915 | 5.853829 | UAGGUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUGG,UAGGAA,UAGGAG,UAGGUG,UAGUUA,UAGUUG |
| KHSRP | 1 | 35 | 0.09090909 | 0.002357873 | 5.268867 | UAGGUA | ACCCUC,AGCCUC,AUAUUU,AUGUAU,AUGUGU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CGGCUU,CUCCCU,CUGCCU,CUGCUU,GCCCUC,GCCUCC,GCCUUC,GGCCUC,UAGGUU,UAUAUU,UAUUUU,UCCCUC,UGCAUG,UGUAUA,UGUGUG,UUAUUA |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.002423369 | 5.229338 | UAGGUA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| ZRANB2 | 1 | 45 | 0.09090909 | 0.003012837 | 4.915230 | AGGUAC | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.007859576 | 3.531901 | UAGGUA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
RBP on BSJ (Exon and Intron)
Plot
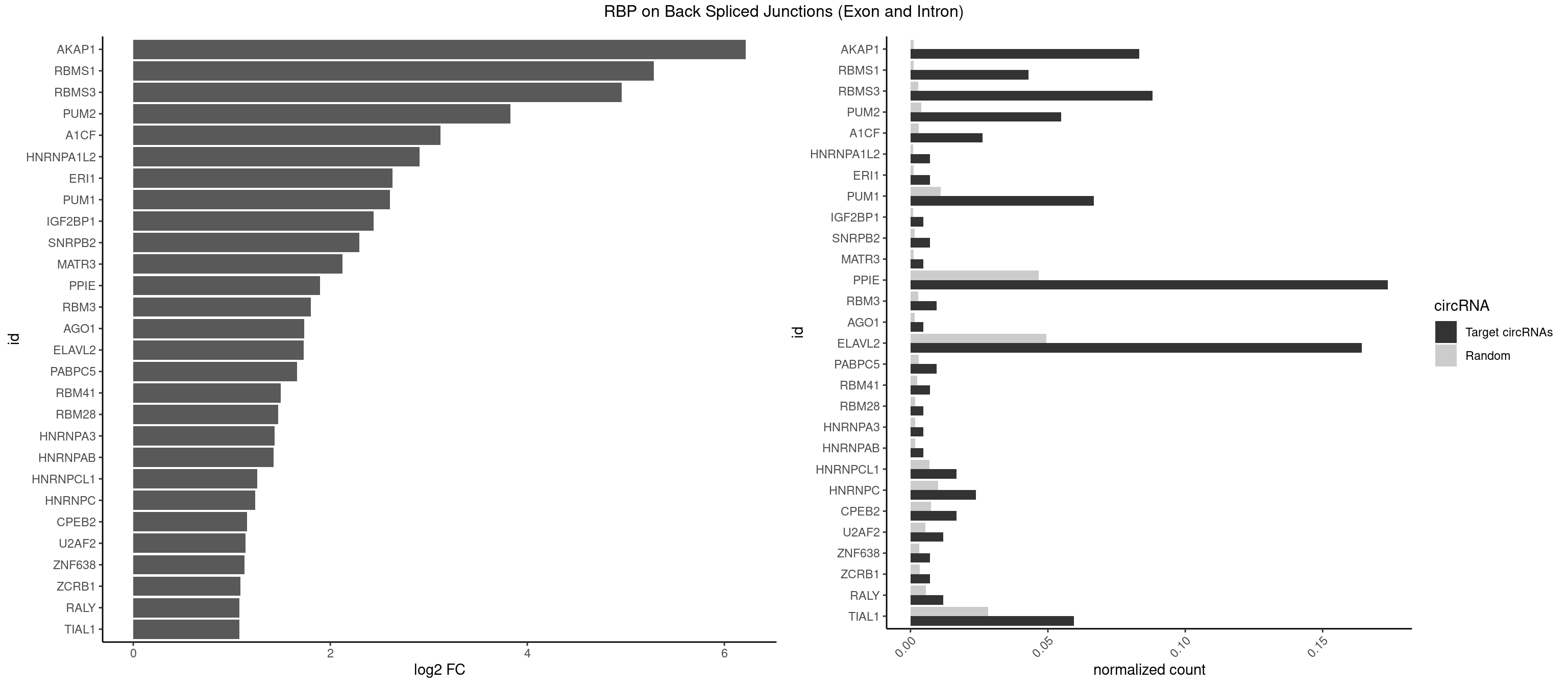
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| AKAP1 | 34 | 221 | 0.083333333 | 0.0011185006 | 6.219256 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| RBMS1 | 17 | 217 | 0.042857143 | 0.0010983474 | 5.286129 | UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBMS3 | 36 | 562 | 0.088095238 | 0.0028365578 | 4.956851 | AAUAUA,AUAUAU,UAUAGA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| PUM2 | 22 | 764 | 0.054761905 | 0.0038542926 | 3.828635 | GUAGAU,GUAUAU,UAGAUA,UAUAUA,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| A1CF | 10 | 598 | 0.026190476 | 0.0030179363 | 3.117408 | AGUAUA,AUAAUU,CAGUAU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| PUM1 | 27 | 2172 | 0.066666667 | 0.0109482064 | 2.606271 | AAUGUU,AAUUGU,ACAUAA,AUUGUA,GUAGAU,GUAUAU,UAGAUA,UAUAUA,UGUAAA,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | GUAUUG,UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| PPIE | 72 | 9262 | 0.173809524 | 0.0466696896 | 1.896949 | AAAUAU,AAUAAU,AAUAUA,AAUUUU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUUAAU,AUUAUU,AUUUAA,AUUUUA,UAAUAA,UAAUUU,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUA,UUAAAA,UUAAAU,UUAAUA,UUAUUU,UUUAAA,UUUAUA,UUUUAA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAACUA,AUACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | AGGUAG | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ELAVL2 | 68 | 9826 | 0.164285714 | 0.0495112858 | 1.730378 | AAUUUG,AAUUUU,AUUAUU,AUUUAA,AUUUUA,AUUUUG,CAUUUU,CUUUCU,CUUUUU,GAUUUU,GUAUUG,UAAUUU,UACUUU,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUGU,UAUUUA,UCUUUU,UGAUUU,UUACUU,UUAGUU,UUAUGU,UUAUUU,UUCUUU,UUGAUU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUCAU,UUUCUU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPCL1 | 6 | 1381 | 0.016666667 | 0.0069629182 | 1.259202 | CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPC | 9 | 2006 | 0.023809524 | 0.0101118501 | 1.235492 | CUUUUU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CPEB2 | 6 | 1487 | 0.016666667 | 0.0074969770 | 1.152585 | CAUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUCUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AUUUAA,GGUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| TIAL1 | 24 | 5597 | 0.059523810 | 0.0282043531 | 1.077549 | AAUUUU,AUUUUA,AUUUUG,CUUUUC,CUUUUU,UUAAAU,UUAUUU,UUUAAA,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.