circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000171863_ENSG00000234171:+:2:3575591:3576630
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000171863_ENSG00000234171:+:2:3575591:3576630 | ENSG00000171863_ENSG00000234171 | MSTRG.17912.8 | + | 2 | 3575592 | 3576630 | 309 | UUCUCCCAGGAGAAAGCCAUGUUCAGUUCGAGCGCCAAGAUCGUGAAGCCCAAUGGCGAGAAGCCGGACGAGUUCGAGUCCGGCAUCUCCCAGGCUCUUCUGGAGCUGGAGAUGAACUCGGACCUCAAGGCUCAGCUCAGGGAGCUGAAUAUUACGGCAGCUAAGGAAAUUGAAGUUGGUGGUGGUCGGAAAGCUAUCAUAAUCUUUGUUCCCGUUCCUCAACUGAAAUCUUUCCAGAAAAUCCAAGUCCGGCUAGUACGCGAAUUGGAGAAAAAGUUCAGUGGGAAGCAUGUCGUCUUUAUCGCUCAGUUCUCCCAGGAGAAAGCCAUGUUCAGUUCGAGCGCCAAGAUCGUGAAGCC | circ |
| ENSG00000171863_ENSG00000234171:+:2:3575591:3576630 | ENSG00000171863_ENSG00000234171 | MSTRG.17912.8 | + | 2 | 3575592 | 3576630 | 22 | UUAUCGCUCAGUUCUCCCAGGA | bsj |
| ENSG00000171863_ENSG00000234171:+:2:3575591:3576630 | ENSG00000171863_ENSG00000234171 | MSTRG.17912.8 | + | 2 | 3575392 | 3575601 | 210 | GGUUGAGGAAAACGCCUUUUGGAGUCAGGCCCUGGAGGGGCGAGCCUUGCUCACAGGGUGGGGAUACAGCCGAUUACCCGCCCUGUGCUUUCCGAUGGCUUCUGCGGGGCGAGCGGGGCCUGGCCGGGGGGUGCGGGCGGGAGGGCGAGCCAGCGGCGCCUGCAGCCCGGGCCGCGUAACGCUGACCGCUGUGCCUUCAGUUCUCCCAGG | ie_up |
| ENSG00000171863_ENSG00000234171:+:2:3575591:3576630 | ENSG00000171863_ENSG00000234171 | MSTRG.17912.8 | + | 2 | 3576621 | 3576830 | 210 | UAUCGCUCAGGUAUCUGUUCUACUGUUGCAGCACGUUUCUGUUUGUGAAUUUUGCUAAAAUUGCUUGUAUUUAGACUGCAUUGGUAGUUGGAGUCAUGAAAACAAUCCUUUUAUGAAUCCAAUUGGGUAGAAAGAUAAAGUACAGGCAGAGCGCCGUGGCUUAGGCCUGUAAUCCCAGCACGGGGAGGUGGAGGCGGGUGGAUCACUGUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
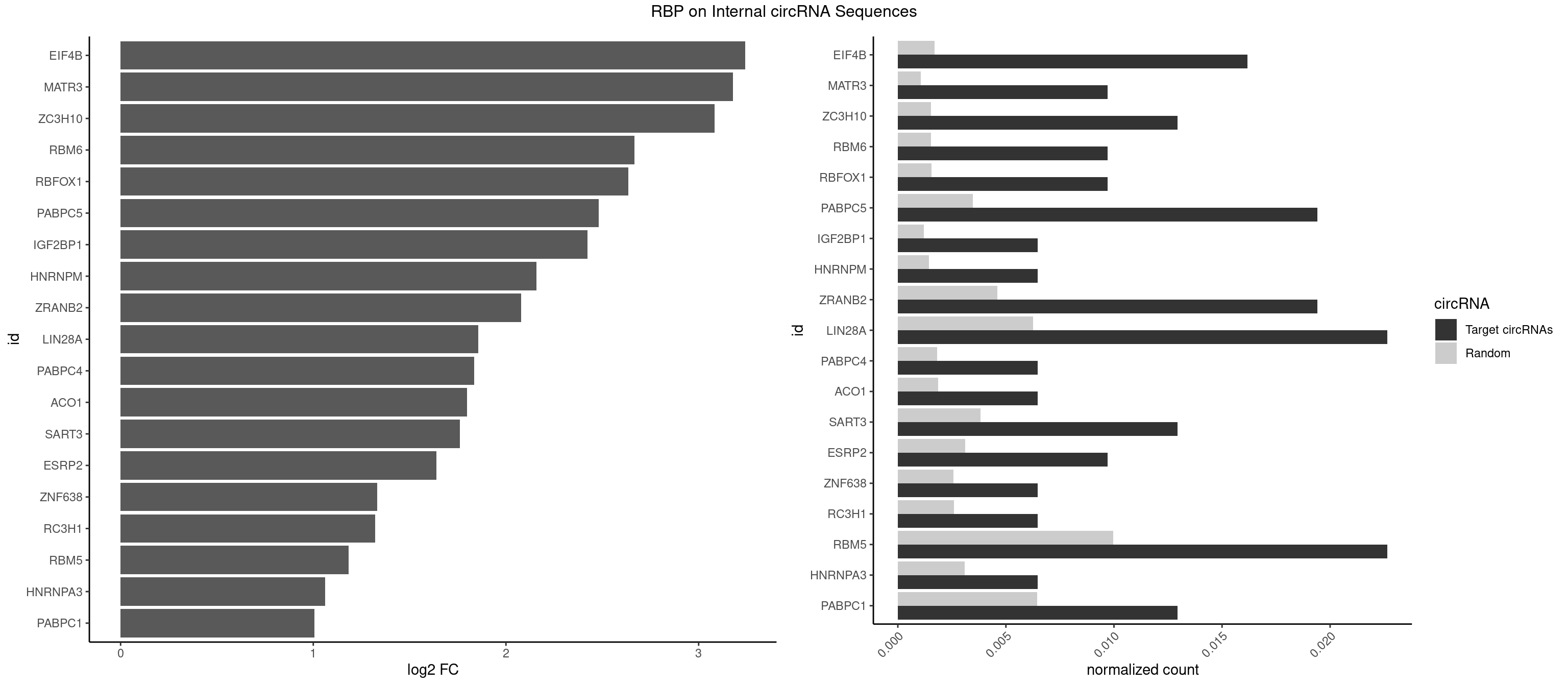
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| EIF4B | 4 | 1179 | 0.016181230 | 0.001710061 | 3.242202 | CUCGGA,GUCGGA,UCGGAA,UCGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| MATR3 | 2 | 739 | 0.009708738 | 0.001072411 | 3.178426 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ZC3H10 | 3 | 1053 | 0.012944984 | 0.001527461 | 3.083186 | CGAGCG,GAGCGC | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBM6 | 2 | 1054 | 0.009708738 | 0.001528910 | 2.666780 | AAUCCA,AUCCAA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBFOX1 | 2 | 1077 | 0.009708738 | 0.001562242 | 2.635666 | AGCAUG,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPC5 | 5 | 2400 | 0.019417476 | 0.003479539 | 2.480388 | AGAAAA,AGAAAG,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP1 | 1 | 831 | 0.006472492 | 0.001205738 | 2.424405 | CCCGUU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPM | 1 | 999 | 0.006472492 | 0.001449204 | 2.159061 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| ZRANB2 | 5 | 3173 | 0.019417476 | 0.004599773 | 2.077721 | GGUGGU,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| LIN28A | 6 | 4315 | 0.022653722 | 0.006254764 | 1.856721 | AGGAGA,GGAGAA,GGAGAU,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| PABPC4 | 1 | 1251 | 0.006472492 | 0.001814403 | 1.834826 | AAAAAG | AAAAAA,AAAAAG |
| ACO1 | 1 | 1283 | 0.006472492 | 0.001860778 | 1.798415 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SART3 | 3 | 2634 | 0.012944984 | 0.003818652 | 1.761258 | AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ESRP2 | 2 | 2150 | 0.009708738 | 0.003117238 | 1.639016 | GGGAAG,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| ZNF638 | 1 | 1773 | 0.006472492 | 0.002570888 | 1.332055 | GUUGGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RC3H1 | 1 | 1786 | 0.006472492 | 0.002589727 | 1.321521 | CUUCUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBM5 | 6 | 6879 | 0.022653722 | 0.009970523 | 1.184007 | AAGGAA,AGGGAG,CUCUUC,GGUGGU,UCUUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| HNRNPA3 | 1 | 2140 | 0.006472492 | 0.003102746 | 1.060776 | GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| PABPC1 | 3 | 4443 | 0.012944984 | 0.006440262 | 1.007202 | AGAAAA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
RBP on BSJ (Exon Only)
Plot
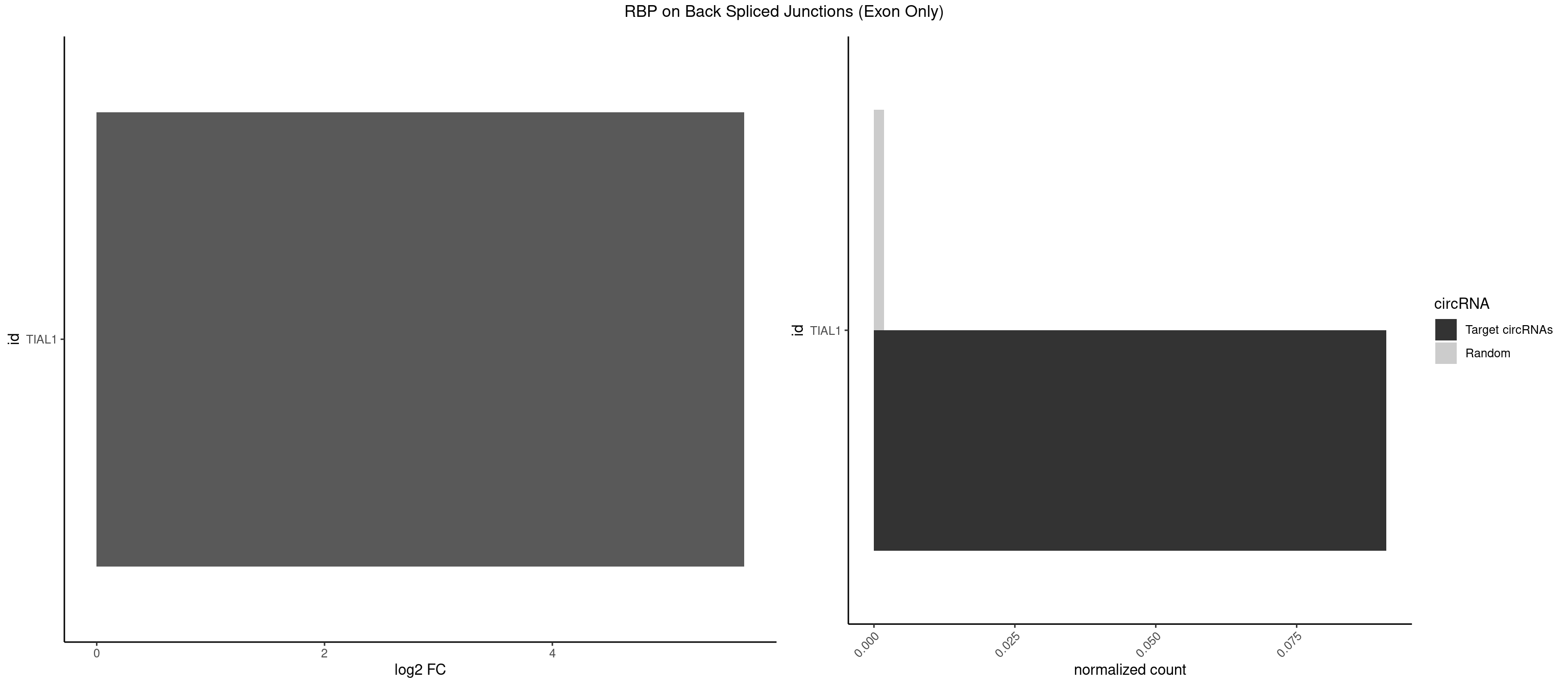
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TIAL1 | 1 | 26 | 0.09090909 | 0.001768405 | 5.683904 | UCAGUU | AAAUUU,AAUUUU,AGUUUU,AUUUUG,CAGUUU,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUCAGU,UUUCAG |
RBP on BSJ (Exon and Intron)
Plot
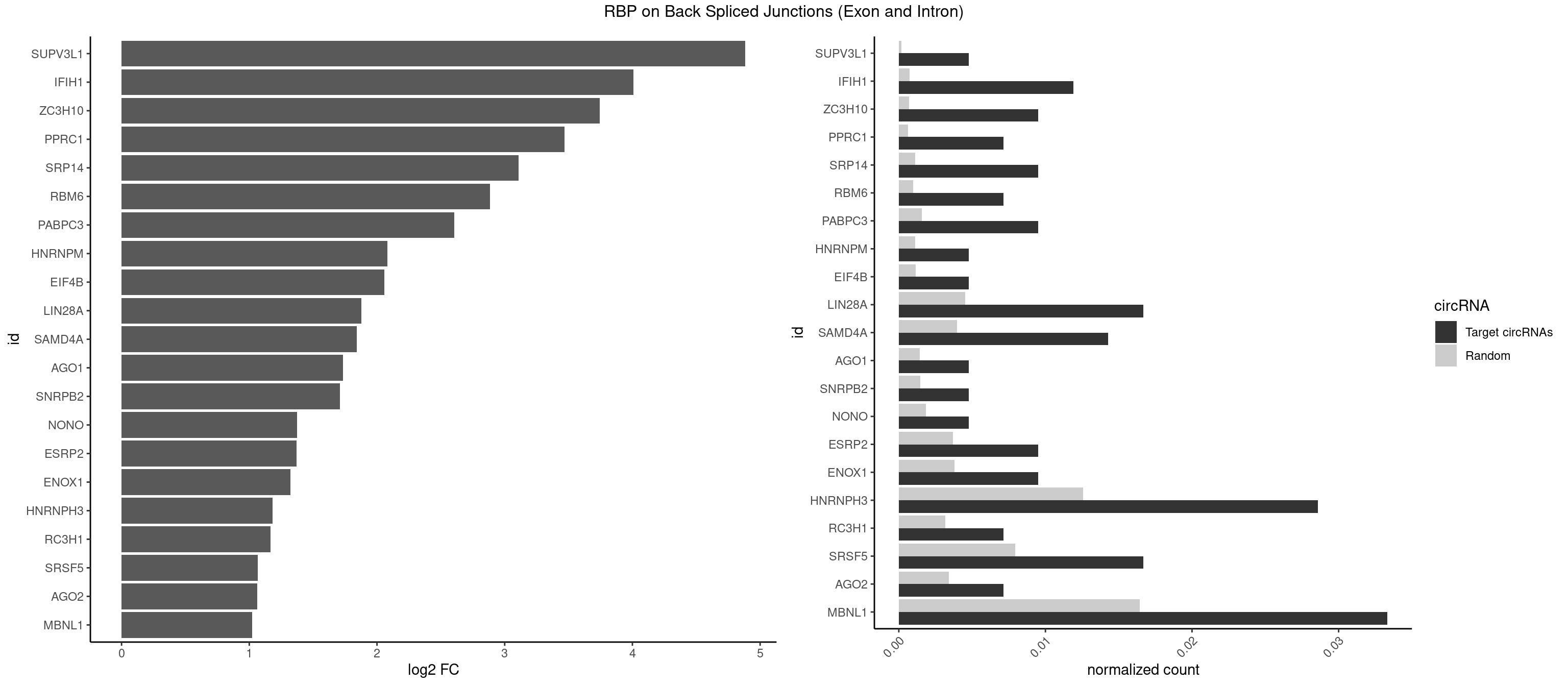
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 1 | 31 | 0.004761905 | 0.0001612253 | 4.884389 | CCGCCC | CCGCCC |
| IFIH1 | 4 | 146 | 0.011904762 | 0.0007406288 | 4.006644 | GCCGCG,GGCCCU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ZC3H10 | 3 | 140 | 0.009523810 | 0.0007103990 | 3.744837 | CCAGCG,CGAGCG,GAGCGC | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PPRC1 | 2 | 127 | 0.007142857 | 0.0006449012 | 3.469351 | CGGCGC,GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| SRP14 | 3 | 218 | 0.009523810 | 0.0011033857 | 3.109602 | CCUGUA,CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA,AUCCAA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| PABPC3 | 3 | 310 | 0.009523810 | 0.0015669085 | 2.603618 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| LIN28A | 6 | 900 | 0.016666667 | 0.0045395002 | 1.876360 | GGAGGG,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SAMD4A | 5 | 790 | 0.014285714 | 0.0039852882 | 1.841817 | CGGGCC,CUGGCC,GCGGGA,GCGGGC,GCGGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | GGUAGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGGAG,GGGGAU,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AGUACA,AUACAG,GUACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPH3 | 11 | 2496 | 0.028571429 | 0.0125806127 | 1.183371 | AUUGGG,CGGGCG,CGGGGG,GAGGGG,GGAGGG,GGGAGG,GGGGAG,GGGGGG,UUGGGU | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CUUCUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SRSF5 | 6 | 1578 | 0.016666667 | 0.0079554615 | 1.066948 | AUAAAG,CGCGUA,GAGGAA,UACAGC,UGCAGC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | GUGCUU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| MBNL1 | 13 | 3258 | 0.033333333 | 0.0164197904 | 1.021530 | ACGCUG,CCGCUG,CGCUGU,CUGCGG,CUUGCU,GCUUGU,GUGCUU,UCGCUC,UGCUUU,UUGCUA,UUGCUC,UUGCUU,UUUGCU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.