circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000131747:-:17:40391505:40411818
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000131747:-:17:40391505:40411818 | ENSG00000131747 | ENST00000423485 | - | 17 | 40391506 | 40411818 | 3478 | GUAAAAGGAUUUCGUAGUUAUGUGGACAUGUAUUUGAAGGACAAGUUGGAUGAAACUGGUAACUCCUUGAAAGUAAUACAUGAACAAGUAAACCACAGGUGGGAAGUGUGUUUAACUAUGAGUGAAAAAGGCUUUCAGCAAAUUAGCUUUGUCAACAGCAUUGCUACAUCCAAGGGUGGCAGACAUGUUGAUUAUGUAGCUGAUCAGAUUGUGACUAAACUUGUUGAUGUUGUGAAGAAGAAGAACAAGGGUGGUGUUGCAGUAAAAGCACAUCAGGUGAAAAAUCACAUGUGGAUUUUUGUAAAUGCCUUAAUUGAAAACCCAACCUUUGACUCUCAGACAAAAGAAAACAUGACUUUACAACCCAAGAGCUUUGGAUCAACAUGCCAAUUGAGUGAAAAAUUUAUCAAAGCUGCCAUUGGCUGUGGUAUUGUAGAAAGCAUACUAAACUGGGUGAAGUUUAAGGCCCAAGUCCAGUUAAACAAGAAGUGUUCAGCUGUAAAACAUAAUAGAAUCAAGGGAAUUCCCAAACUCGAUGAUGCCAAUGAUGCAGGGGGCCGAAACUCCACUGAGUGUACGCUUAUCCUGACUGAGGGAGAUUCAGCCAAAACUUUGGCUGUUUCAGGCCUUGGUGUGGUUGGGAGAGACAAAUAUGGGGUUUUCCCUCUUAGAGGAAAAAUACUCAAUGUUCGAGAAGCUUCUCAUAAGCAGAUCAUGGAAAAUGCUGAGAUUAACAAUAUCAUCAAGAUUGUGGGUCUUCAGUACAAGAAAAACUAUGAAGAUGAAGAUUCAUUGAAGACGCUUCGUUAUGGGAAGAUAAUGAUUAUGACAGAUCAGGACCAAGAUGGUUCCCACAUCAAAGGCUUGCUGAUUAAUUUUAUCCAUCACAACUGGCCCUCUCUUCUGCGACAUCGUUUUCUGGAGGAAUUUAUCACUCCCAUUGUAAAGGUAUCUAAAAACAAGCAAGAAAUGGCAUUUUACAGCCUUCCUGAAUUUGAAGAGUGGAAGAGUUCUACUCCAAAUCAUAAAAAAUGGAAAGUCAAAUAUUACAAAGGUUUGGGCACCAGCACAUCAAAGGAAGCUAAAGAAUACUUUGCAGAUAUGAAAAGACAUCGUAUCCAGUUCAAAUAUUCUGGUCCUGAAGAUGAUGCUGCUAUCAGCCUGGCCUUUAGCAAAAAACAGAUAGAUGAUCGAAAGGAAUGGUUAACUAAUUUCAUGGAGGAUAGAAGACAACGAAAGUUACUUGGGCUUCCUGAGGAUUACUUGUAUGGACAAACUACCACAUAUCUGACAUAUAAUGACUUCAUCAACAAGGAACUUAUCUUGUUCUCAAAUUCUGAUAACGAGAGAUCUAUCCCUUCUAUGGUGGAUGGUUUGAAACCAGGUCAGAGAAAGGUUUUGUUUACUUGCUUCAAACGGAAUGACAAGCGAGAAGUAAAGGUUGCCCAAUUAGCUGGAUCAGUGGCUGAAAUGUCUUCUUAUCAUCAUGGUGAGAUGUCACUAAUGAUGACCAUUAUCAAUUUGGCUCAGAAUUUUGUGGGUAGCAAUAAUCUAAACCUCUUGCAGCCCAUUGGUCAGUUUGGUACCAGGCUACAUGGUGGCAAGGAUUCUGCUAGUCCACGAUACAUCUUUACAAUGCUCAGCUCUUUGGCUCGAUUGUUAUUUCCACCAAAAGAUGAUCACACGUUGAAGUUUUUAUAUGAUGACAACCAGCGUGUUGAGCCUGAAUGGUACAUUCCUAUUAUUCCCAUGGUGCUGAUAAAUGGUGCUGAAGGAAUCGGUACUGGGUGGUCCUGCAAAAUCCCCAACUUUGAUGUGCGUGAAAUUGUAAAUAACAUCAGGCGUUUGAUGGAUGGAGAAGAACCUUUGCCAAUGCUUCCAAGUUACAAGAACUUCAAGGGUACUAUUGAAGAACUGGCUCCAAAUCAAUAUGUGAUUAGUGGUGAAGUAGCUAUUCUUAAUUCUACAACCAUUGAAAUCUCAGAGCUUCCCGUCAGAACAUGGACCCAGACAUACAAAGAACAAGUUCUAGAACCCAUGUUGAAUGGCACCGAGAAGACACCUCCUCUCAUAACAGACUAUAGGGAAUACCAUACAGAUACCACUGUGAAAUUUGUUGUGAAGAUGACUGAAGAAAAACUGGCAGAGGCAGAGAGAGUUGGACUACACAAAGUCUUCAAACUCCAAACUAGUCUCACAUGCAACUCUAUGGUGCUUUUUGACCACGUAGGCUGUUUAAAGAAAUAUGACACGGUGUUGGAUAUUCUAAGAGACUUUUUUGAACUCAGACUUAAAUAUUAUGGAUUAAGAAAAGAAUGGCUCCUAGGAAUGCUUGGUGCUGAAUCUGCUAAACUGAAUAAUCAGGCUCGCUUUAUCUUAGAGAAAAUAGAUGGCAAAAUAAUCAUUGAAAAUAAGCCUAAGAAAGAAUUAAUUAAAGUUCUGAUUCAGAGGGGAUAUGAUUCGGAUCCUGUGAAGGCCUGGAAAGAAGCCCAGCAAAAGGUUCCAGAUGAAGAAGAAAAUGAAGAGAGUGACAACGAAAAGGAAACUGAAAAGAGUGACUCCGUAACAGAUUCUGGACCAACCUUCAACUAUCUUCUUGAUAUGCCCCUUUGGUAUUUAACCAAGGAAAAGAAAGAUGAACUCUGCAGGCUAAGAAAUGAAAAAGAACAAGAGCUGGACACAUUAAAAAGAAAGAGUCCAUCAGAUUUGUGGAAAGAAGACUUGGCUACAUUUAUUGAAGAAUUGGAGGCUGUUGAAGCCAAGGAAAAACAAGAUGAACAAGUCGGACUUCCUGGGAAAGGGGGGAAGGCCAAGGGGAAAAAAACACAAAUGGCUGAAGUUUUGCCUUCUCCGCGUGGUCAAAGAGUCAUUCCACGAAUAACCAUAGAAAUGAAAGCAGAGGCAGAAAAGAAAAAUAAAAAGAAAAUUAAGAAUGAAAAUACUGAAGGAAGCCCUCAAGAAGAUGGUGUGGAACUAGAAGGCCUAAAACAAAGAUUAGAAAAGAAACAGAAAAGAGAACCAGGUACAAAGACAAAGAAACAAACUACAUUGGCAUUUAAGCCAAUCAAAAAAGGAAAGAAGAGAAAUCCCUGGUCUGAUUCAGAAUCAGAUAGGAGCAGUGACGAAAGUAAUUUUGAUGUCCCUCCACGAGAAACAGAGCCACGGAGAGCAGCAACAAAAACAAAAUUCACAAUGGAUUUGGAUUCAGAUGAAGAUUUCUCAGAUUUUGAUGAAAAAACUGAUGAUGAAGAUUUUGUCCCAUCAGAUGCUAGUCCACCUAAGACCAAAACUUCCCCAAAACUUAGUAACAAAGAACUGAAACCACAGAAAAGUGUCGUGUCAGACCUUGAAGCUGAUGAUGUUAAGGGCAGUGUACCACUGUCUUCAAGCCCUCCUGCUACACAUUUCCCAGAUGAAACUGAAAUUACAAACCCAGUUCCUAAAAAGAAUGUGACAGUGAAGAAGACAGCAGCAAAAAGUAAAAGGAUUUCGUAGUUAUGUGGACAUGUAUUUGAAGGACAAGUUGGA | circ |
| ENSG00000131747:-:17:40391505:40411818 | ENSG00000131747 | ENST00000423485 | - | 17 | 40391506 | 40411818 | 22 | AGCAGCAAAAAGUAAAAGGAUU | bsj |
| ENSG00000131747:-:17:40391505:40411818 | ENSG00000131747 | ENST00000423485 | - | 17 | 40411809 | 40412018 | 210 | GAUCUCACUAUGUUGCCCAGGGUGGUCUUGAACUCCUGGGCUCAAGUGAUCCACCCACCUCGGCCUGUGUCCUUUAAUGACCAUUCCCUUAUGCCUAUCAGUGAACAUCAUUGCAUUGGUUUUGGAAAGUCCUCAUAGUCUAUCAUUGAACCUAUUUUUUAAUAACUUUCUUAAUACUGUUACCUUUAAUUCCUGUACAGGUAAAAGGAU | ie_up |
| ENSG00000131747:-:17:40391505:40411818 | ENSG00000131747 | ENST00000423485 | - | 17 | 40391306 | 40391515 | 210 | GCAGCAAAAAGUAAGCCUAAAUCUUUGAGAUGGGUUAAUGUUGCAAUUACCUAACUGGUUUCCACGUGUCUAUUUCAAUUUUUUUAUUGCCAAAACUUACUAUUGAUAUUACAGAUUAAAUGUUUUCCAAUUGGAAGCAAUUUCUUUUCGAUCUUUAUAAUCAAAAUUAGUAGUCAAGGCUGUUCCAAAACAGUAAGUUAUCUCUAUUGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
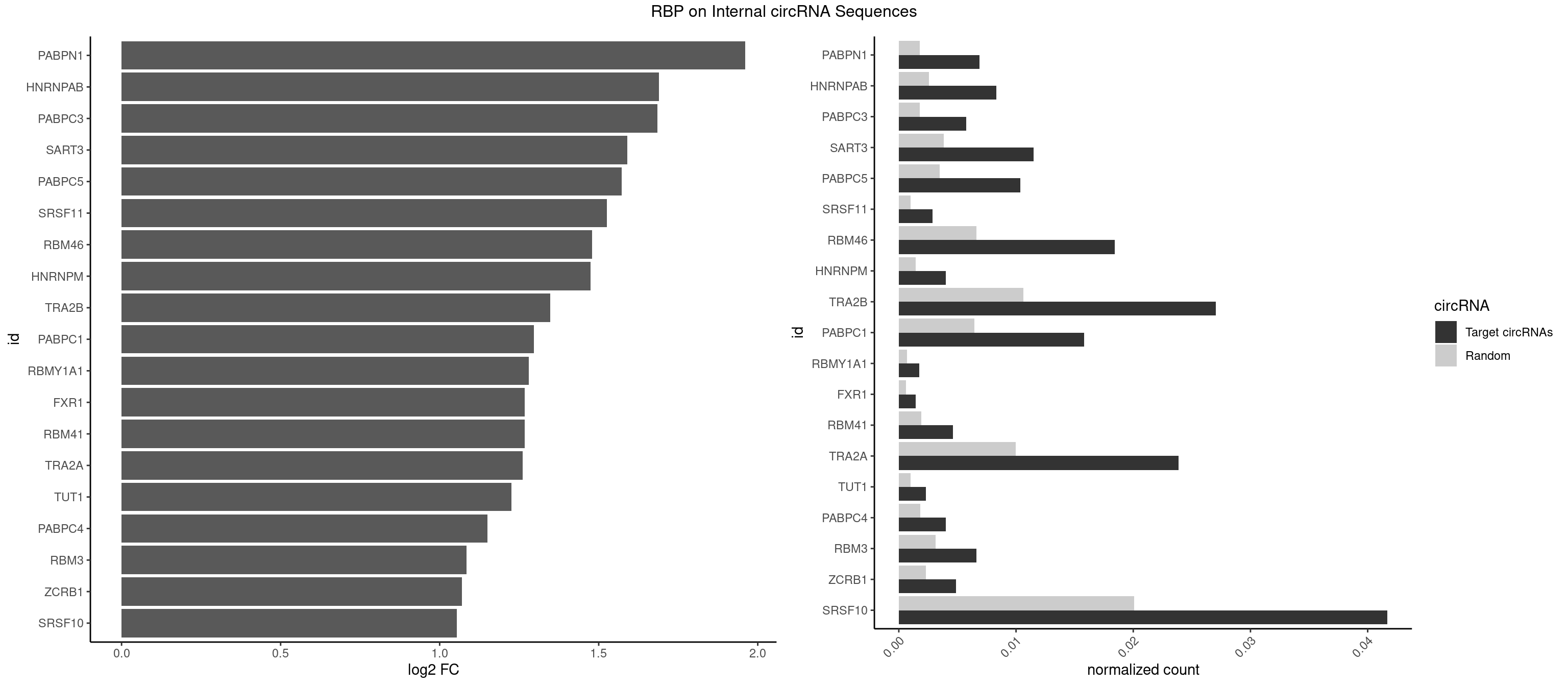
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PABPN1 | 23 | 1222 | 0.006900518 | 0.0017723764 | 1.961020 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| HNRNPAB | 28 | 1782 | 0.008338125 | 0.0025839306 | 1.690156 | AAAGAC,AAGACA,ACAAAG,AGACAA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPC3 | 19 | 1234 | 0.005750431 | 0.0017897669 | 1.683898 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SART3 | 39 | 2634 | 0.011500863 | 0.0038186524 | 1.590607 | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPC5 | 35 | 2400 | 0.010350776 | 0.0034795387 | 1.572771 | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SRSF11 | 9 | 688 | 0.002875216 | 0.0009985015 | 1.525834 | AAGAAG | AAGAAG |
| RBM46 | 63 | 4554 | 0.018401380 | 0.0066011240 | 1.479030 | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPM | 13 | 999 | 0.004025302 | 0.0014492040 | 1.473836 | AAGGAA,GAAGGA,GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| TRA2B | 93 | 7329 | 0.027027027 | 0.0106226650 | 1.347257 | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| PABPC1 | 54 | 4443 | 0.015813686 | 0.0064402624 | 1.295982 | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAU,ACUAAU,AGAAAA,CAAACC,CAAAUA,CAAAUC,CGAAUA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| RBMY1A1 | 5 | 489 | 0.001725129 | 0.0007101099 | 1.280590 | ACAAGA | ACAAGA,CAAGAC |
| FXR1 | 4 | 411 | 0.001437608 | 0.0005970720 | 1.267693 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM41 | 15 | 1318 | 0.004600345 | 0.0019115000 | 1.267037 | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| TRA2A | 82 | 6871 | 0.023864290 | 0.0099589296 | 1.260791 | AAAGAA,AAGAAA,AAGAAG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| TUT1 | 7 | 678 | 0.002300173 | 0.0009840095 | 1.224998 | AAAUAC,AAUACU,AGAUAC,CGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC4 | 13 | 1251 | 0.004025302 | 0.0018144033 | 1.149602 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBM3 | 22 | 2152 | 0.006612996 | 0.0031201361 | 1.083695 | AAAACU,AAACUA,AAGACG,AAGACU,AAUACU,AGACUA,AUACUA,GAAACU,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ZCRB1 | 16 | 1605 | 0.004887867 | 0.0023274215 | 1.070472 | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GGAUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SRSF10 | 144 | 13860 | 0.041690627 | 0.0200874160 | 1.053431 | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGAGA,GAGAGC,GAGGAA,GAGGGA,GAGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
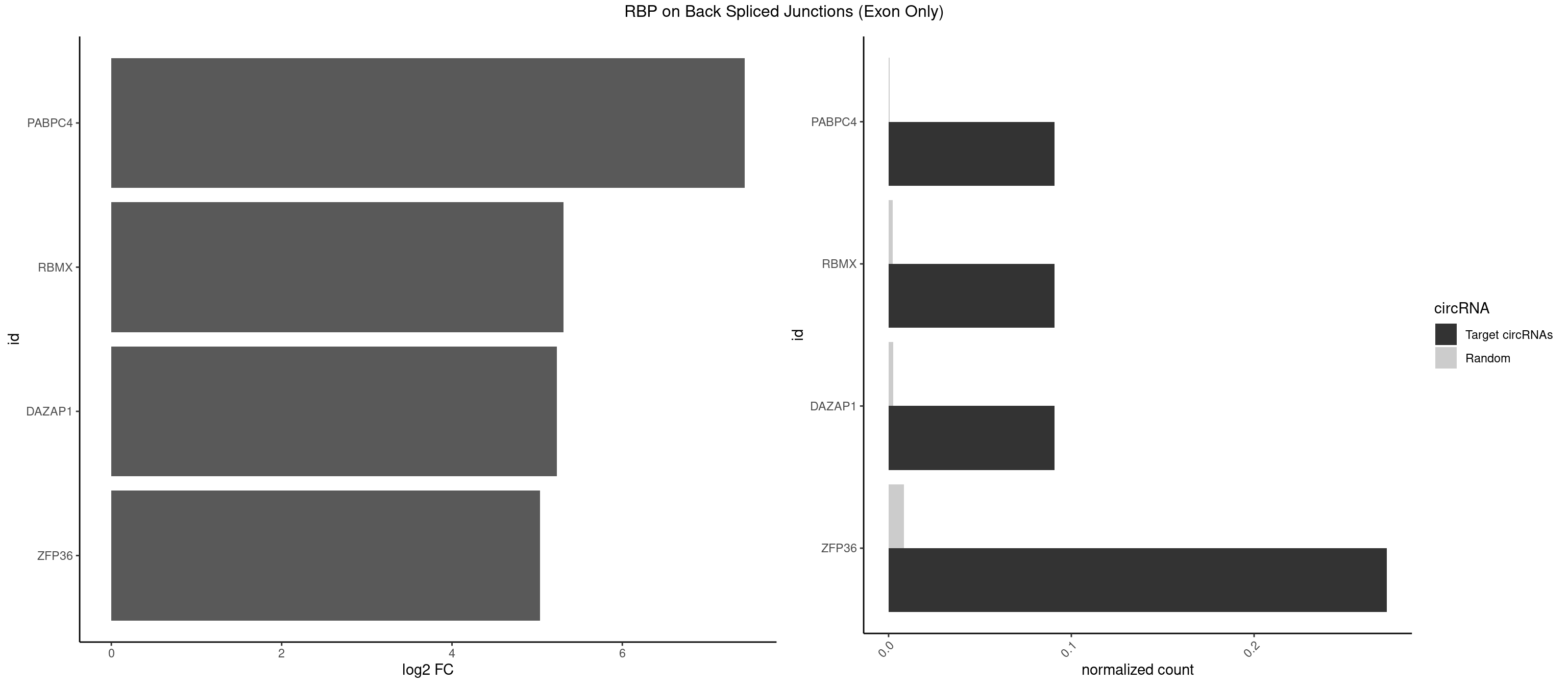
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PABPC4 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | AAAAAG | AAAAAA,AAAAAG |
| RBMX | 1 | 34 | 0.09090909 | 0.0022923762 | 5.309509 | AAGUAA | AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUGUU,AUCAAA,AUCCCA,GAAGGA,GGAAGG,UAAGAC,UCAAAA |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.0024233691 | 5.229338 | AGUAAA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| ZFP36 | 5 | 126 | 0.27272727 | 0.0083180508 | 5.035070 | AAAAAG,AAAAGU,AAAGUA,AAGUAA,AGUAAA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
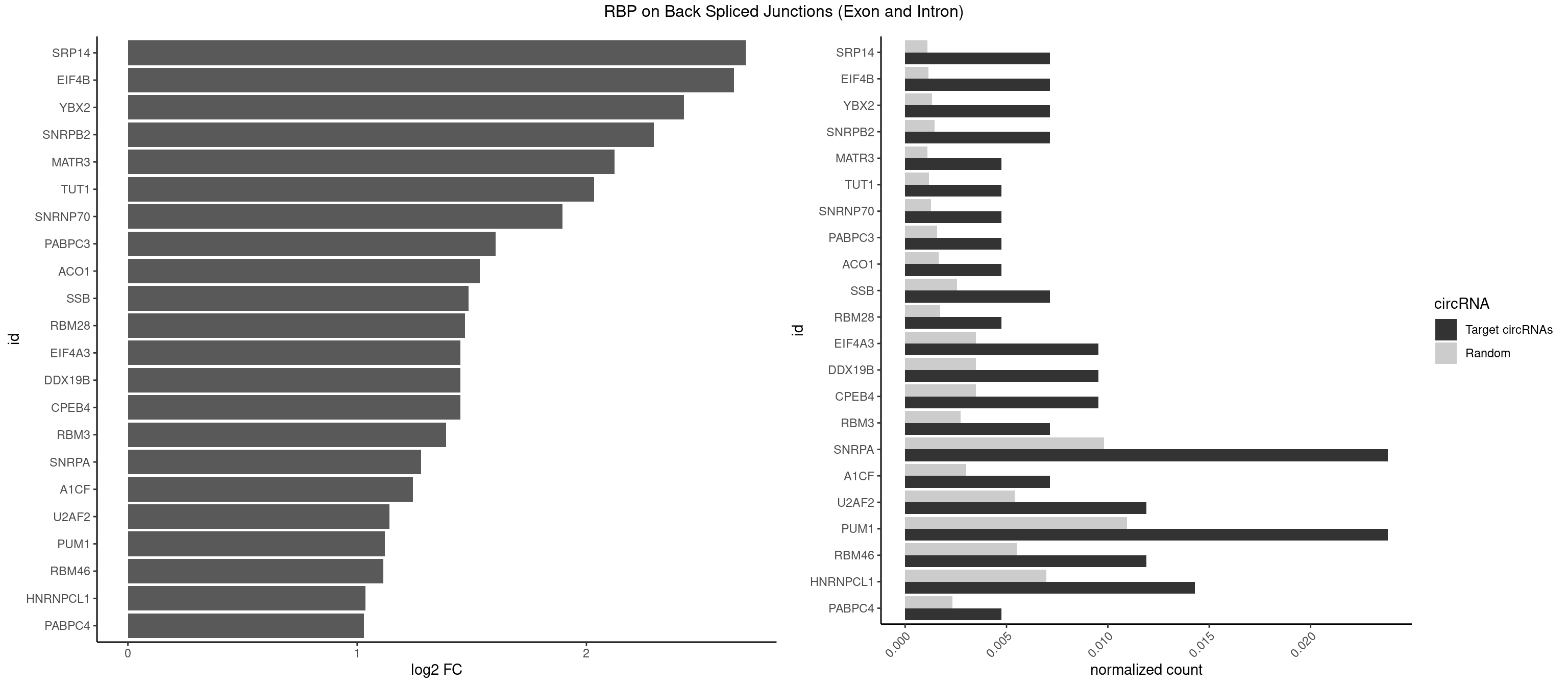
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRP14 | 2 | 218 | 0.007142857 | 0.001103386 | 2.694564 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| EIF4B | 2 | 226 | 0.007142857 | 0.001143692 | 2.642803 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| YBX2 | 2 | 263 | 0.007142857 | 0.001330109 | 2.424957 | AACAUC,ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.001456066 | 2.294425 | AUUGCA,UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| MATR3 | 1 | 216 | 0.004761905 | 0.001093309 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| TUT1 | 1 | 230 | 0.004761905 | 0.001163845 | 2.032640 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.001279726 | 1.895704 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.001566909 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ACO1 | 1 | 325 | 0.004761905 | 0.001642483 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SSB | 2 | 505 | 0.007142857 | 0.002549375 | 1.486358 | GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.001718057 | 1.470761 | AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| CPEB4 | 3 | 691 | 0.009523810 | 0.003486497 | 1.449760 | UUUUUU | UUUUUU |
| DDX19B | 3 | 691 | 0.009523810 | 0.003486497 | 1.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 3 | 691 | 0.009523810 | 0.003486497 | 1.449760 | UUUUUU | UUUUUU |
| RBM3 | 2 | 541 | 0.007142857 | 0.002730754 | 1.387202 | AAAACU,AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SNRPA | 9 | 1947 | 0.023809524 | 0.009814591 | 1.278539 | AGUAGU,AUUCCU,AUUGCA,GUAGUC,GUUACC,GUUUCC,UUACCU,UUCCUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| A1CF | 2 | 598 | 0.007142857 | 0.003017936 | 1.242939 | AUCAGU,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.005401048 | 1.140228 | UUUUCC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| PUM1 | 9 | 2172 | 0.023809524 | 0.010948206 | 1.120844 | AAUGUU,UAAUGU,UGUACA,UUAAUG,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.005501814 | 1.113560 | AAUCAA,AUCAAA,AUCAUU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.006962918 | 1.036809 | AUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.002332729 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.