circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000037637:-:1:16294782:16315435
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000037637:-:1:16294782:16315435 | ENSG00000037637 | ENST00000375592 | - | 1 | 16294783 | 16315435 | 519 | CUGUUGAGUGGAAUGUCAUGGCCAGCUCCUCGGACAGUGAAGAUGACAGUUUCAUGGCUGUGGACCAGGAAGAAACUGUGCUGGAAGGGACAAUGGAUCAAGAUGAGGAGCCCCACCCAGUAUUGGAGGCUGAGGAGACUAGACAUAAUAGGUCCAUGUCGGAGCUGCCAGAAGAGGUUUUGGAGUAUAUCCUGUCCUUUCUCUCACCGUAUCAGGAACACAAAACUGCGGCCCUUGUCUGCAAACAGUGGUAUCGACUUAUCAAAGGUGUAGCCCAUCAGUGUUAUCAUGGUUUCAUGAAGGCUGUCCAGGAAGGAAACAUUCAGUGGGAGAGCCGUACCUAUCCUUAUCCUGGAACCCCAAUCACUCAGCGCUUCUCGCACAGUGCAUGCUAUUAUGAUGCUAAUCAGUCUAUGUAUGUGUUUGGAGGCUGUACCCAGAGCAGCUGCAAUGCUGCUUUCAAUGACCUCUGGAGACUUGACCUAAACAGCAAAGAGUGGAUCCGACCUUUGGCUUCAGCUGUUGAGUGGAAUGUCAUGGCCAGCUCCUCGGACAGUGAAGAUGACAGU | circ |
| ENSG00000037637:-:1:16294782:16315435 | ENSG00000037637 | ENST00000375592 | - | 1 | 16294783 | 16315435 | 22 | UUUGGCUUCAGCUGUUGAGUGG | bsj |
| ENSG00000037637:-:1:16294782:16315435 | ENSG00000037637 | ENST00000375592 | - | 1 | 16315426 | 16315635 | 210 | AAAAGCCUUUGGGGAUGGGCUUGGUAUUUGAGCUGUACCUGUCGAAUUAAGGUGGCUCACAAGUGGUAUUGGAAAGUGCUGAAGAGCAGAUUUCACACUGAGAGUGGAAACGAAAUUACAAAGAGCUUGGAUGUACAUGCCUGAAUUGUUUUUGAGCUGCUUUCAGUGGAAUACUGAUAUUUAUGUUUUACCUAUUACAGCUGUUGAGUG | ie_up |
| ENSG00000037637:-:1:16294782:16315435 | ENSG00000037637 | ENST00000375592 | - | 1 | 16294583 | 16294792 | 210 | UUGGCUUCAGGUAAGAGAUGAAAUGCUGAUCUUUGCCUCCUUACCAGGUGACUCCCUUGGCAUGGAAACUCUACCUGAUCAUUUUUGGUGUUGGUCUCAAGGAUGGGUUACUGGGACUCAGGUCCUUAAGUUUAAUGUGCCAUGUAAAUGAUAAUAGUAAUAUGCUUGUGUAAGUCUUUGUUUUUUGUUAAAUUAUACUUUAUCAUUAGU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
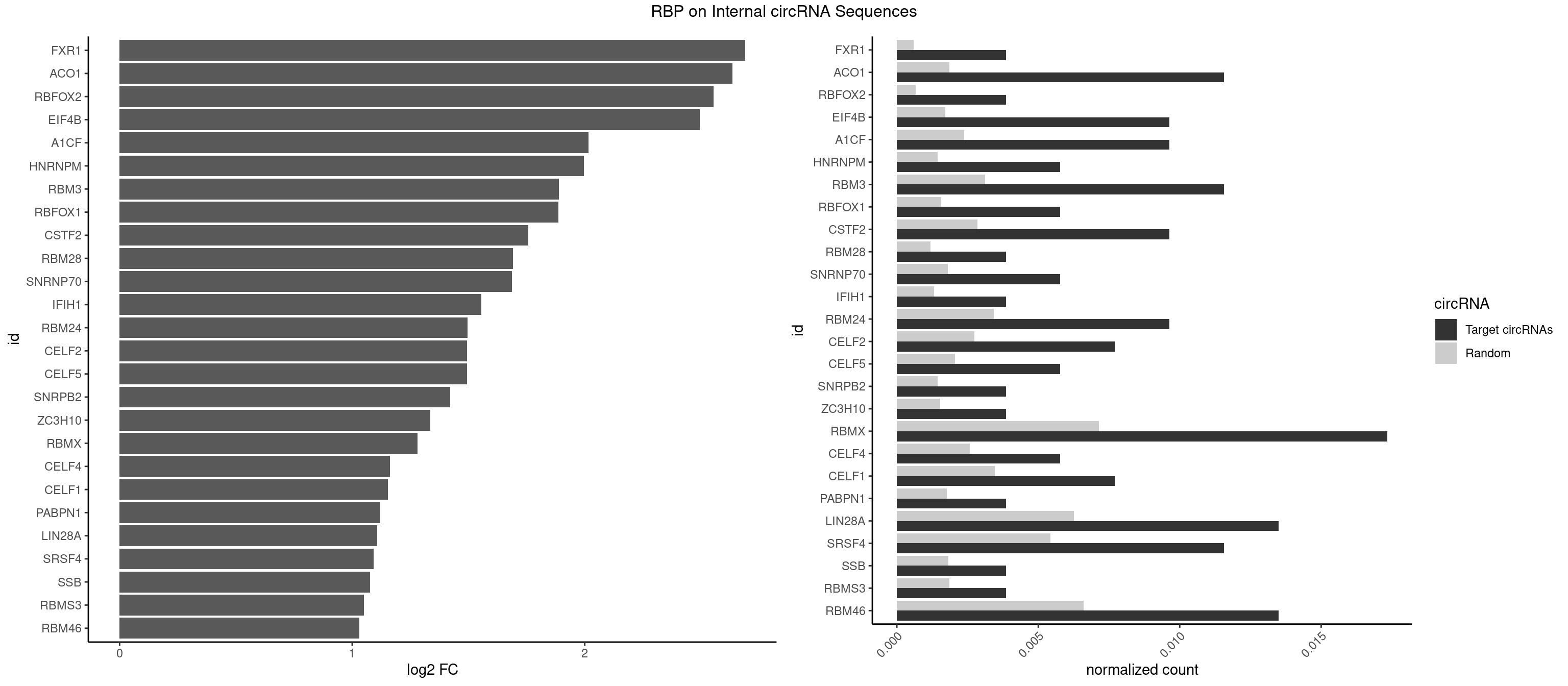
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 411 | 0.003853565 | 0.0005970720 | 2.690217 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| ACO1 | 5 | 1283 | 0.011560694 | 0.0018607779 | 2.635250 | CAGUGA,CAGUGC,CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBFOX2 | 1 | 452 | 0.003853565 | 0.0006564894 | 2.553350 | UGCAUG | UGACUG,UGCAUG |
| EIF4B | 4 | 1179 | 0.009633911 | 0.0017100607 | 2.494074 | CUCGGA,GUCGGA,UCGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| A1CF | 4 | 1642 | 0.009633911 | 0.0023810421 | 2.016529 | AGUAUA,AUCAGU,CAGUAU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPM | 2 | 999 | 0.005780347 | 0.0014492040 | 1.995895 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM3 | 5 | 2152 | 0.011560694 | 0.0031201361 | 1.889547 | AAAACU,AGACUA,GAAACU,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBFOX1 | 2 | 1077 | 0.005780347 | 0.0015622419 | 1.887538 | GCAUGC,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| CSTF2 | 4 | 1967 | 0.009633911 | 0.0028520334 | 1.756131 | GUGUUU,GUUUUG,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBM28 | 1 | 822 | 0.003853565 | 0.0011926949 | 1.691969 | GUGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SNRNP70 | 2 | 1237 | 0.005780347 | 0.0017941145 | 1.687884 | AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| IFIH1 | 1 | 904 | 0.003853565 | 0.0013115296 | 1.554943 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM24 | 4 | 2357 | 0.009633911 | 0.0034172229 | 1.495297 | AGAGUG,GAGUGG,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF2 | 3 | 1886 | 0.007707129 | 0.0027346479 | 1.494838 | AUGUGU,GUAUGU,UAUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 2 | 1415 | 0.005780347 | 0.0020520728 | 1.494074 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SNRPB2 | 1 | 991 | 0.003853565 | 0.0014376103 | 1.422521 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ZC3H10 | 1 | 1053 | 0.003853565 | 0.0015274610 | 1.335058 | CAGCGC | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBMX | 8 | 4925 | 0.017341040 | 0.0071387787 | 1.280441 | AAGGAA,AGGAAG,AGUGUU,AUCAAA,GAAGGA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| CELF4 | 2 | 1782 | 0.005780347 | 0.0025839306 | 1.161589 | GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF1 | 3 | 2391 | 0.007707129 | 0.0034664959 | 1.152716 | UGUCUG,UGUGUU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| PABPN1 | 1 | 1222 | 0.003853565 | 0.0017723764 | 1.120509 | AGAAGA | AAAAGA,AGAAGA |
| LIN28A | 6 | 4315 | 0.013487476 | 0.0062547643 | 1.108593 | AGGAGA,GGAGUA,UGGAGA,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SRSF4 | 5 | 3740 | 0.011560694 | 0.0054214720 | 1.092471 | AGAAGA,AGGAAG,GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| SSB | 1 | 1260 | 0.003853565 | 0.0018274462 | 1.076365 | GCUGUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBMS3 | 1 | 1283 | 0.003853565 | 0.0018607779 | 1.050288 | UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBM46 | 6 | 4554 | 0.013487476 | 0.0066011240 | 1.030837 | AUCAAA,AUCAAG,AUCAUG,AUGAAG,AUGAUG,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
RBP on BSJ (Exon Only)
Plot
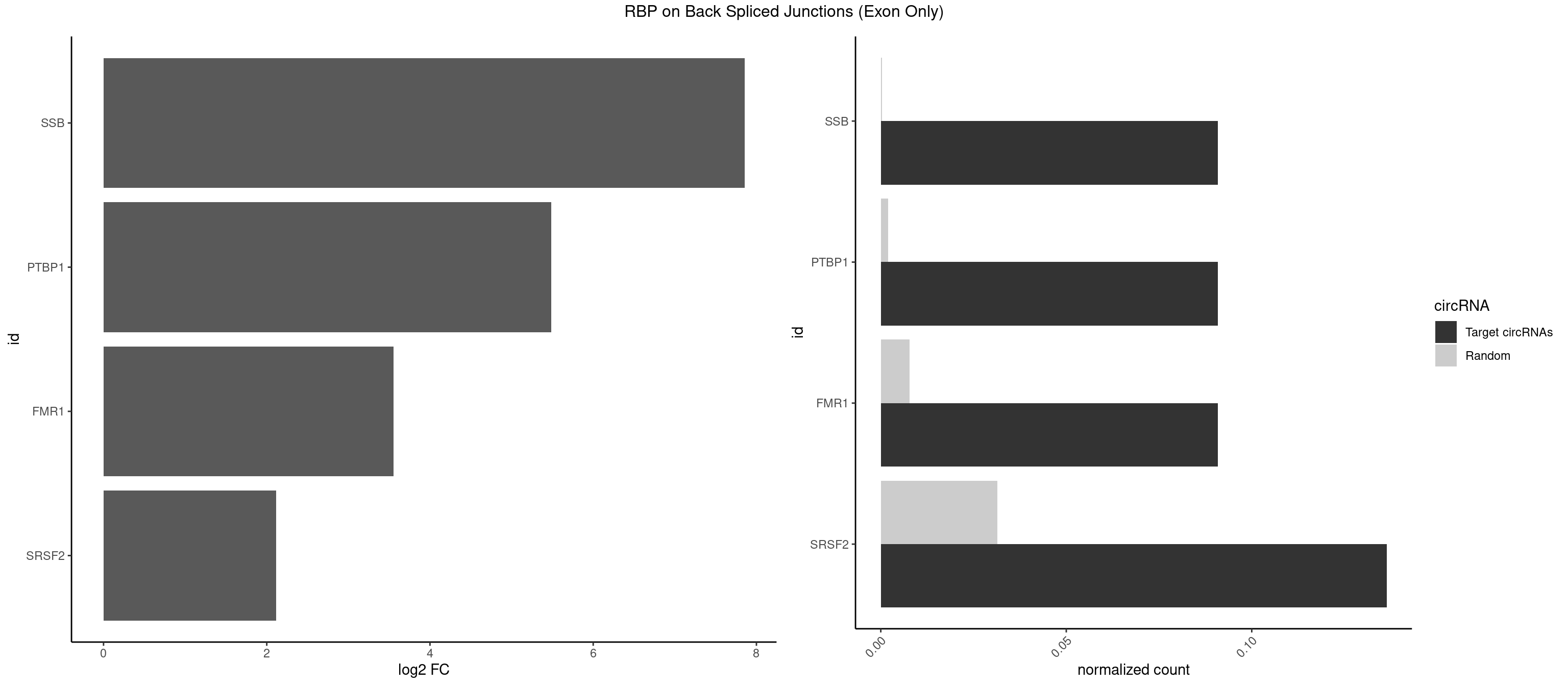
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SSB | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GCUGUU | CUGUUU,GCUGUU,UGCUGU |
| PTBP1 | 1 | 30 | 0.09090909 | 0.0020303904 | 5.484596 | AGCUGU | AGCUGU,CAUCUU,CCAUCU,CCUCUU,CUAUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUUCU,GGCUCC,GUCUUU,UACUUU,UAUUUU,UCCUCU,UCUAUC,UCUCUU,UCUUCU,UCUUUC,UUACUU,UUCUUC,UUCUUG,UUUCUU |
| FMR1 | 1 | 117 | 0.09090909 | 0.0077285827 | 3.556149 | CAGCUG | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF2 | 2 | 479 | 0.13636364 | 0.0314383023 | 2.116864 | AGCUGU,GCUGUU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
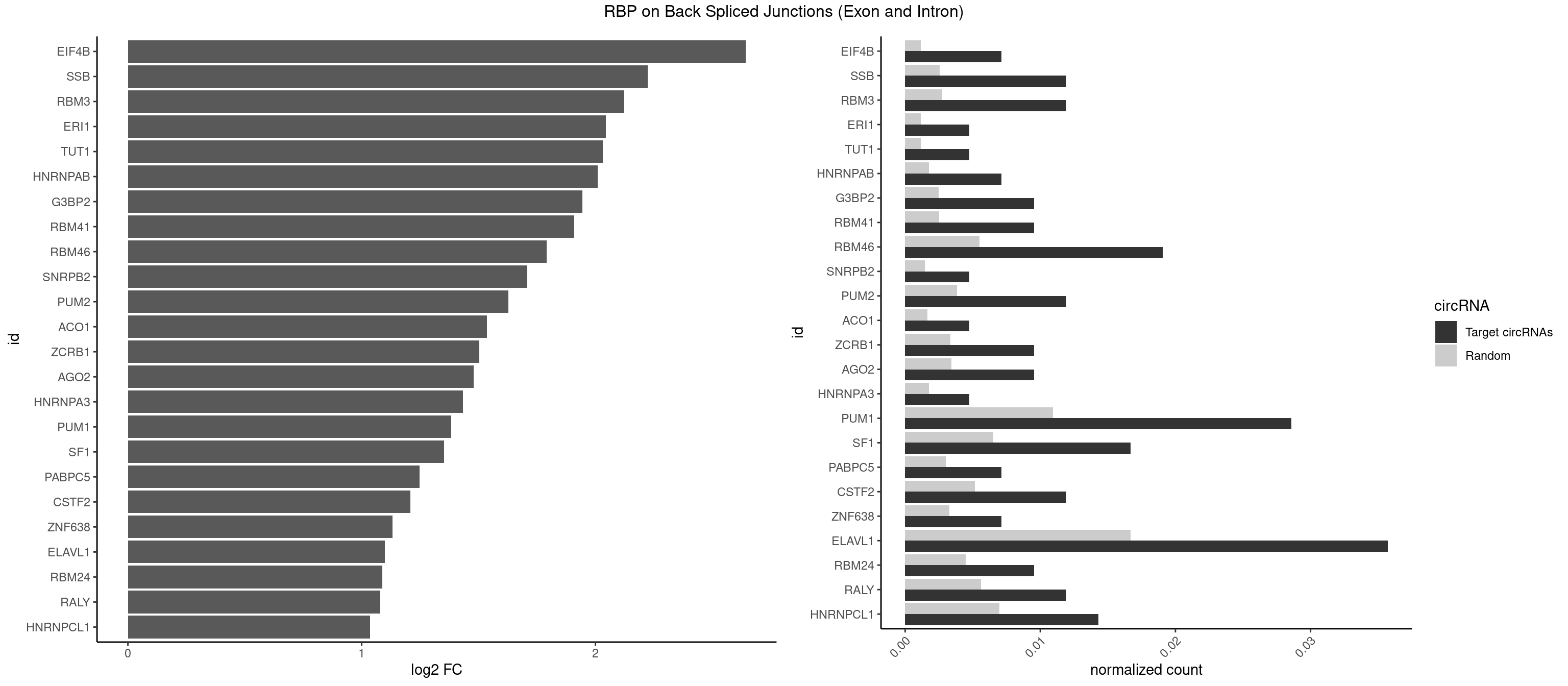
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| EIF4B | 2 | 226 | 0.007142857 | 0.001143692 | 2.642803 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SSB | 4 | 505 | 0.011904762 | 0.002549375 | 2.223323 | GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM3 | 4 | 541 | 0.011904762 | 0.002730754 | 2.124168 | AAACGA,AAUACU,GAAACG,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ERI1 | 1 | 228 | 0.004761905 | 0.001153769 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 1 | 230 | 0.004761905 | 0.001163845 | 2.032640 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.001773478 | 2.009919 | ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| G3BP2 | 3 | 490 | 0.009523810 | 0.002473801 | 1.944809 | AGGAUG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM41 | 3 | 502 | 0.009523810 | 0.002534260 | 1.909974 | AUACUU,UACAUG,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM46 | 7 | 1091 | 0.019047619 | 0.005501814 | 1.791631 | AAUGAU,AUCAUU,AUGAAA,AUGAUA,GAUCAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.001456066 | 1.709463 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PUM2 | 4 | 764 | 0.011904762 | 0.003854293 | 1.627001 | GUAAAU,GUACAU,UGUAAA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ACO1 | 1 | 325 | 0.004761905 | 0.001642483 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.003360540 | 1.502846 | AAUUAA,GAAUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| AGO2 | 3 | 677 | 0.009523810 | 0.003415961 | 1.479247 | AAAGUG,AAGUGC,AGUGCU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.001763402 | 1.433177 | CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| PUM1 | 11 | 2172 | 0.028571429 | 0.010948206 | 1.383879 | AAUUGU,GAAUUG,GUAAAU,GUAAUA,GUACAU,UAAUAU,UAAUGU,UGUAAA,UGUACA,UUAAUG,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| SF1 | 6 | 1294 | 0.016666667 | 0.006524587 | 1.353007 | CACUGA,UACUGA,UAGUAA,UAUACU,UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| PABPC5 | 2 | 596 | 0.007142857 | 0.003007860 | 1.247764 | GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.005154172 | 1.207726 | GUGUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.003259774 | 1.131729 | GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ELAVL1 | 14 | 3309 | 0.035714286 | 0.016676743 | 1.098664 | AUUUAU,UAUUUA,UGUUUU,UUGUUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RBM24 | 3 | 888 | 0.009523810 | 0.004479041 | 1.088349 | AGAGUG,GAGUGG,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| RALY | 4 | 1117 | 0.011904762 | 0.005632809 | 1.079612 | UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.006962918 | 1.036809 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.