circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000176715:+:16:89098590:89133262
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000176715:+:16:89098590:89133262 | ENSG00000176715 | MSTRG.13252.9 | + | 16 | 89098591 | 89133262 | 1559 | GUGUCCCACCGGCCUUCCGGGUUCCAGCGCCAGGCCUGGUGCCUGCCCCAGGAGGAUGAGCAUUUGCAUUGCCUGCACCUUAUCGUGCCCUUCCACCUGCUGAAGCAGCUGUGCCUGCCGCUCUUGUGAACUGCGAACUUCCCCUUACCUCCUCUCUCUGGCUCGGGAGCUGGCUCGGCCGCCUGUCAGUGCAAUGCUGCCCCAUGUGGUGCUCACCUUCCGGCGCCUGGGCUGCGCCUUGGCGUCCUGCCGGCUGGCGCCUGCGAGACACAGAGGAAGUGGUCUUCUGCACACAGCCCCAGUGGCCCGCUCGGACAGGAGCGCCCCGGUGUUCACCCGUGCCCUGGCCUUUGGGGACAGAAUCGCCCUGGUUGACCAGCACGGCCGCCACACGUACAGGGAGCUUUAUUCCCGCAGCCUUCGCCUGUCCCAGGAGAUCUGCAGGCUCUGCGGGUGUGUCGGCGGGGACCUCCGGGAGGAGAGGGUCUCCUUCCUAUGCGCUAACGAUGCCUCCUACGUCGUGGCCCAGUGGGCGUCAUGGAUGAGUGGCGGUGUGGCAGUCCCCCUCUACAGGAAGCAUCCCGCGGCCCAGCUGGAGUAUGUCAUCUGCGACUCCCAGAGCUCUGUGGUCCUUGCCAGCCAGGAGUACCUGGAGCUCCUGAGCCCGGUGGUCAGGAAGCUGGGGGUCCCGCUGCUGCCGCUCACACCAGCCAUCUACACUGGAGCAGUAGAGGAACCGGCAGAGGUCCCGGUCCCAGAGCAGGGAUGGAGGAACAAGGGCGCCAUGAUCAUCUACACCAGUGGGACCACGGGGAGGCCCAAGGGCGUGCUGAGCACGCACCAAAACAUCAGGGCUGUGGUGACCGGGCUGGUCCACAAGUGGGCAUGGACCAAAGACGACGUGAUCCUCCACGUGCUCCCGCUGCACCACGUCCAUGGUGUGGUCAACGCGCUGCUCUGUCCUCUCUGGGUGGGAGCCACCUGUGUGAUGAUGCCUGAGUUCAGCCCUCAGCAGGUUUGGGAAAAGUUCUUAAGUUCUGAAACGCCGCGGAUCAAUGUCUUUAUGGCAGUGCCUACAAUAUACACCAAGCUGAUGGAGUACUACGACAGGCAUUUUACCCAGCCGCACGCCCAGGAUUUCUUGCGUGCAGUUUGUGAAGAAAAAAUUAGGCUGAUGGUCUCAGGCUCAGCUGCCCUGCCCCUCCCAGUGCUGGAGAAGUGGAAGAACAUCACGGGCCACACCCUGCUGGAGCGGUAUGGCAUGACCGAGAUCGGCAUGGCUCUGUCCGGGCCCCUGACCACUGCCGUGCGCCUGCCAGGUUCCGUGGGGACCCCACUGCCUGGAGUACAGGUGCGCAUUGUCUCAGAAAACCCACAGAGGGAAGCCUGCUCCUACACCAUCCACGCAGAGGGAGACGAGAGGGGGACCAAGGUGACCCCAGGGUUUGAAGAAAAGGAGGGGGAGCUGCUGGUGAGGGGACCCUCCGUGUUUCGAGAAUACUGGAAUAAACCAGAAGAAACUAAGAGUGCAUUCACCCUGGAUGGCUGGUUUAAGACAGGUGUCCCACCGGCCUUCCGGGUUCCAGCGCCAGGCCUGGUGCCUGCCCCA | circ |
| ENSG00000176715:+:16:89098590:89133262 | ENSG00000176715 | MSTRG.13252.9 | + | 16 | 89098591 | 89133262 | 22 | GUUUAAGACAGGUGUCCCACCG | bsj |
| ENSG00000176715:+:16:89098590:89133262 | ENSG00000176715 | MSTRG.13252.9 | + | 16 | 89098391 | 89098600 | 210 | GUGUGUGGGAGGAGACUCGACAGGGUCAUGGGGUCACUGCUGCCCGUUGUCGGGAGAUAAGGCUCACAGCUUUAUUUGAUAAUGUUGUCUUUAGCUUGUAAAUGAACAUGCUUCCCUUCCCCCAUUGAGUGUUGAGUGUUGUGCCUGUGGGAAGCGUUAUACACACAGCCUGGGACCUCAGCACAGAUGCCCGUUGACAGGUGUCCCACC | ie_up |
| ENSG00000176715:+:16:89098590:89133262 | ENSG00000176715 | MSTRG.13252.9 | + | 16 | 89133253 | 89133462 | 210 | UUUAAGACAGGUAGGACCCAGCCCCAUGGGAGUGGAGGAGACCCCGAGGGGACAGGCAGGAACUCAUUGCUGCCCACGUUGAGUGACACCGAGGCUGGGAGUUCCCAGAAUUUUCAGCCAAAGGCAGAAGAUGGGGUGGAGUGGGGCUGGGGGCCACUGUUACGGCACUGCCUCCUGAGCACCCGGCCUCCCCACCGGCUUCUAGAGUGG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
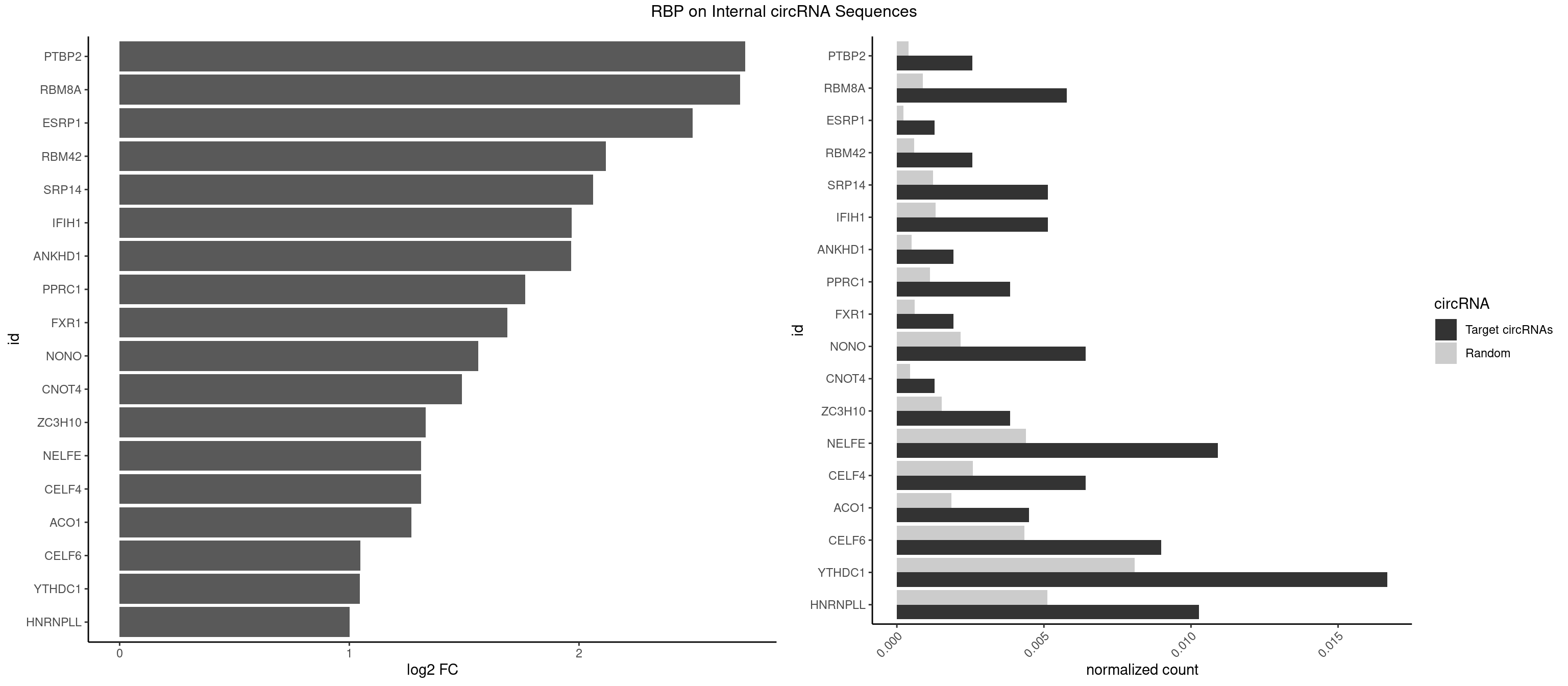
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 3 | 267 | 0.002565747 | 0.0003883867 | 2.723814 | CUCUCU | CUCUCU |
| RBM8A | 8 | 611 | 0.005772931 | 0.0008869128 | 2.702440 | ACGCGC,AUGCGC,CGCGCU,GUGCGC,UGCGCC,UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| ESRP1 | 1 | 156 | 0.001282874 | 0.0002275250 | 2.495282 | AGGGAU | AGGGAU |
| RBM42 | 3 | 407 | 0.002565747 | 0.0005912752 | 2.117477 | AACUAA,ACUAAG,ACUACG | AACUAA,AACUAC,ACUAAG,ACUACG |
| SRP14 | 7 | 847 | 0.005131495 | 0.0012289250 | 2.061982 | CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 | 7 | 904 | 0.005131495 | 0.0013115296 | 1.968129 | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCGC | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ANKHD1 | 2 | 339 | 0.001924310 | 0.0004927293 | 1.965474 | AGACGA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| PPRC1 | 5 | 780 | 0.003848621 | 0.0011318283 | 1.765686 | CGGCGC,GGCGCC,GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| FXR1 | 2 | 411 | 0.001924310 | 0.0005970720 | 1.688365 | ACGACA,ACGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| NONO | 9 | 1498 | 0.006414368 | 0.0021723567 | 1.562046 | AGAGGA,AGGAAC,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| CNOT4 | 1 | 314 | 0.001282874 | 0.0004564992 | 1.490695 | GACAGA | GACAGA |
| ZC3H10 | 5 | 1053 | 0.003848621 | 0.0015274610 | 1.333206 | CAGCGC,CCAGCG,GAGCGC,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| NELFE | 16 | 3028 | 0.010904426 | 0.0043896388 | 1.312740 | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGUCUC,UCUCUC,UCUCUG,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| CELF4 | 9 | 1782 | 0.006414368 | 0.0025839306 | 1.311740 | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ACO1 | 6 | 1283 | 0.004490058 | 0.0018607779 | 1.270828 | CAGUGC,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CELF6 | 13 | 2997 | 0.008980115 | 0.0043447134 | 1.047473 | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,UGUGAU,UGUGGU,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| YTHDC1 | 25 | 5576 | 0.016677357 | 0.0080822104 | 1.045069 | GAAUAC,GAGUAC,GAGUGC,GCCUAC,GCCUGC,GCGUGC,GGCUGC,UACUAC,UCCUAC,UCCUGC,UCGUGC,UGAUGC,UGCUGC,UGGUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| HNRNPLL | 15 | 3534 | 0.010262989 | 0.0051229360 | 1.002408 | ACACCA,ACGACA,AGACGA,CACACA,CACACC,CACCAC,CACUGC,GACGAC,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
RBP on BSJ (Exon Only)
Plot
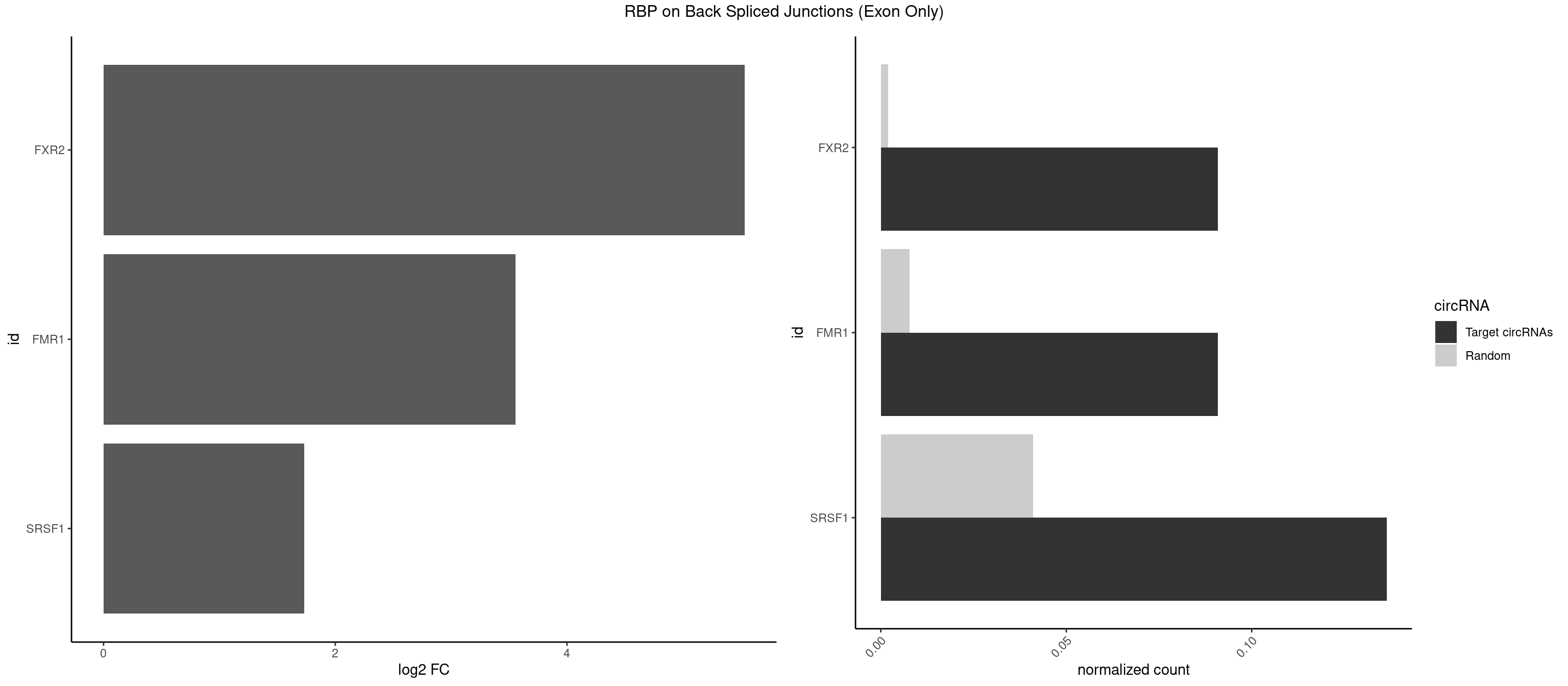
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR2 | 1 | 29 | 0.09090909 | 0.001964894 | 5.531901 | GACAGG | AGACAA,AGACAG,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGGA,GGACAA,GGACAG,GGACGA,UGACAA,UGACAG,UGACGA |
| FMR1 | 1 | 117 | 0.09090909 | 0.007728583 | 3.556149 | GACAGG | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF1 | 2 | 625 | 0.13636364 | 0.041000786 | 1.733736 | ACAGGU,GACAGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
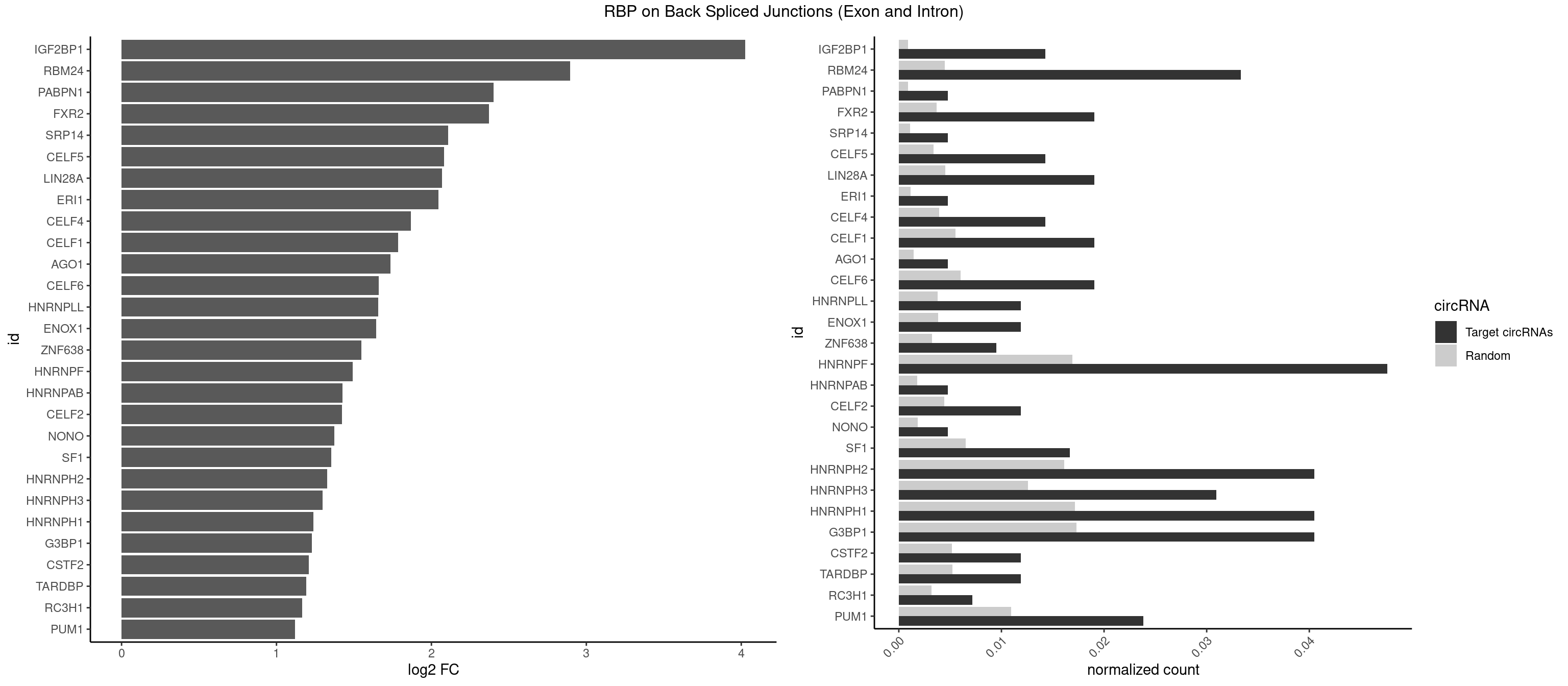
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGF2BP1 | 5 | 173 | 0.014285714 | 0.0008766626 | 4.026408 | AGCACC,CACCCG,CCCGUU,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| RBM24 | 13 | 888 | 0.033333333 | 0.0044790407 | 2.895704 | AGAGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGG,GUGUGU,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| FXR2 | 7 | 730 | 0.019047619 | 0.0036829907 | 2.370661 | AGACAG,GACAGG,GGACAG,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| CELF5 | 5 | 669 | 0.014285714 | 0.0033756550 | 2.081334 | GUGUGG,GUGUGU,GUGUUG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| LIN28A | 7 | 900 | 0.019047619 | 0.0045395002 | 2.069005 | AGGAGA,GGAGAU,GGAGGA,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| CELF4 | 5 | 776 | 0.014285714 | 0.0039147521 | 1.867580 | GUGUGG,GUGUGU,GUGUUG,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF1 | 7 | 1097 | 0.019047619 | 0.0055320435 | 1.783726 | GUGUGU,GUGUUG,GUUGUG,UGUGUG,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | AGGUAG | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| CELF6 | 7 | 1196 | 0.019047619 | 0.0060308343 | 1.659181 | GUGGGG,GUGUGG,GUGUUG,UGUGGG,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | ACACAC,CACACA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| ENOX1 | 4 | 756 | 0.011904762 | 0.0038139863 | 1.642167 | AAGACA,AGACAG,GGACAG,UAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | CGUUGU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPF | 19 | 3360 | 0.047619048 | 0.0169336961 | 1.491642 | AGGGGA,AUGGGA,AUGGGG,CUGGGG,GAGGGG,GAUGGG,GGAGGA,GGGAAG,GGGAGG,GGGCUG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CELF2 | 4 | 881 | 0.011904762 | 0.0044437727 | 1.421682 | GUGUGU,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SF1 | 6 | 1294 | 0.016666667 | 0.0065245869 | 1.353007 | CACAGA,CACCGA,GCUGCC,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| HNRNPH2 | 16 | 3198 | 0.040476190 | 0.0161174929 | 1.328446 | AGGGGA,CGAGGG,CUGGGG,GAGGGG,GGAGGA,GGGAAG,GGGAGG,GGGCUG,GGGGCU,GGGGGC,UGGGGG,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH3 | 12 | 2496 | 0.030952381 | 0.0125806127 | 1.298848 | AGGGGA,CGAGGG,GAGGGG,GGAGGA,GGGAAG,GGGAGG,GGGCUG,GGGGCU,GGGGGC,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 16 | 3406 | 0.040476190 | 0.0171654575 | 1.237565 | AGGGGA,CGAGGG,CUGGGG,GAGGGG,GGAGGA,GGGAAG,GGGAGG,GGGCUG,GGGGCU,GGGGGC,UGGGGG,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| G3BP1 | 16 | 3431 | 0.040476190 | 0.0172914148 | 1.227018 | ACAGGC,ACCGGC,AGGCAG,CACCCG,CACCGG,CAGGCA,CCACCG,CCCACC,CCCACG,CCCCAC,CCCCCA,CCCGGC,CCGGCC,CGGCAC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUGU,GUGUUG,UGUGUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUGUGU,GUUGUG,UGUGUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CCCUUC,UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PUM1 | 9 | 2172 | 0.023809524 | 0.0109482064 | 1.120844 | AAUGUU,AGAAUU,AGAUAA,CAGAAU,CCAGAA,CUUGUA,GUAAAU,UAAUGU,UGUAAA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.