circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000107560:-:10:118009081:118015110
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000107560:-:10:118009081:118015110 | ENSG00000107560 | MSTRG.4703.4 | - | 10 | 118009082 | 118015110 | 190 | UUUUCAUAUGAGUCCAACAAGCAAUGAAGACCUCAGGAAAAUCCCGGACAGCAACCCCUUUGAUGCCACUGCAGGGUAUCGUAGUCUGACCUAUGAAGAGGUUCUACAGGAGCUGGUGAAACACAAAGAACUCCUUAGGAGGAAAGACACCCACAUCCGGGAACUCGAGGACUACAUCGACAACCUCCUUUUUUCAUAUGAGUCCAACAAGCAAUGAAGACCUCAGGAAAAUCCCGGACA | circ |
| ENSG00000107560:-:10:118009081:118015110 | ENSG00000107560 | MSTRG.4703.4 | - | 10 | 118009082 | 118015110 | 22 | ACAACCUCCUUUUUUCAUAUGA | bsj |
| ENSG00000107560:-:10:118009081:118015110 | ENSG00000107560 | MSTRG.4703.4 | - | 10 | 118015101 | 118015310 | 210 | CAAAGUUUGAAAUUGUAAUGAUGCUUUAAGAAUGUUGGGGAAAUACUGGAUUUUUUUCUACUUCUUGGAUUAAAUUUCACUCCUCUGAAUAAUGUAGGGCUUUAGAAAUUUGUAUUACAAUGUUGGUUAAAAUUUUUCUAUUUUUAUGUUUCCUCCCACAUGAGUUAUGACACUUUUAACAUUUGUUUGUGUUUUAUUAGUUUUCAUAUG | ie_up |
| ENSG00000107560:-:10:118009081:118015110 | ENSG00000107560 | MSTRG.4703.4 | - | 10 | 118008882 | 118009091 | 210 | CAACCUCCUUGUAAGGGUAAUGGAAGAAACGCCCAGUAUUCUCAGAGUGCCGUAUGAACCAUCCAGGAAAGCUGGCAAAUUCUCUAACAGUUAAUAAAGCCAAUUGUAUUGUAUUGAUAAUUGGACAAAGAGAGAGAAAGAAAGAAGGAAGGAAGGAAAAACUUGUUACUGAAAGAGACUACACUAUCAGGUUUCAUUACAUAUUCUCCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
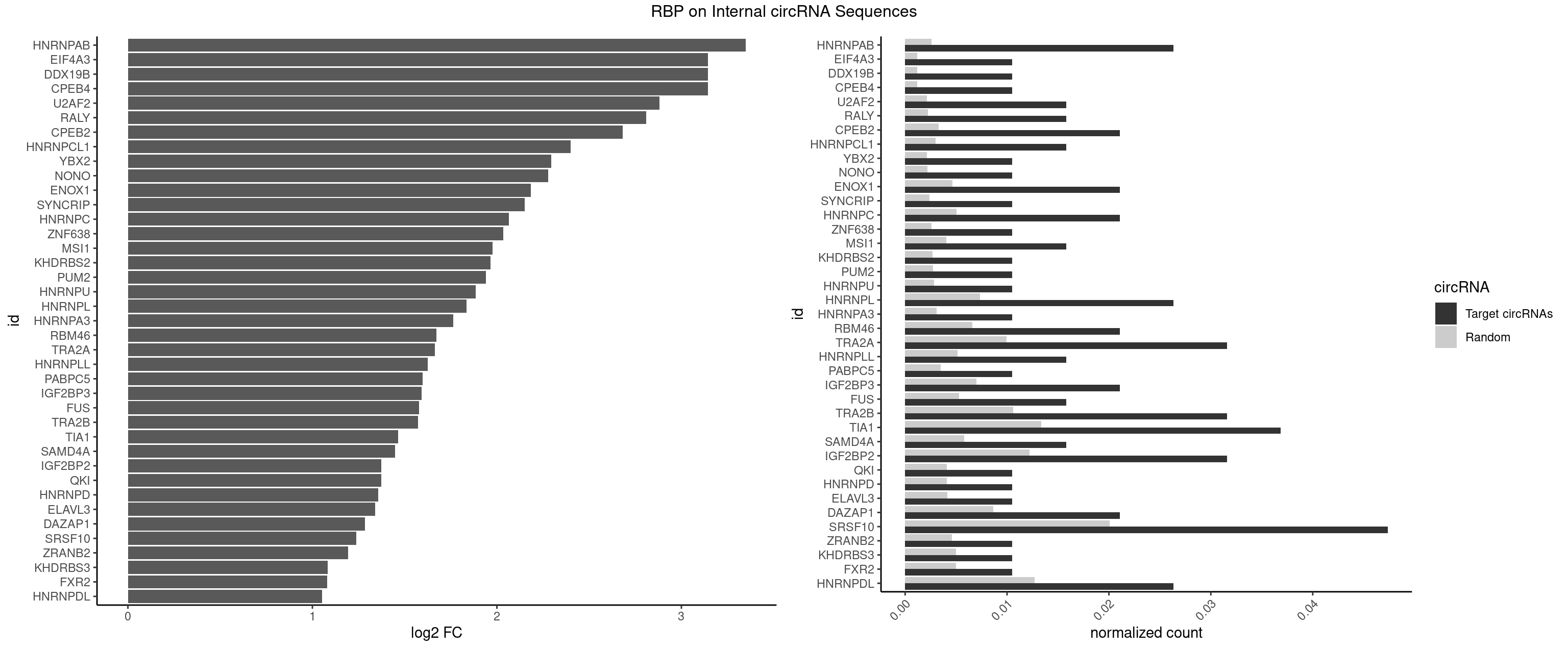
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPAB | 4 | 1782 | 0.02631579 | 0.002583931 | 3.348289 | AAAGAC,AAGACA,ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CPEB4 | 1 | 821 | 0.01052632 | 0.001191246 | 3.143458 | UUUUUU | UUUUUU |
| DDX19B | 1 | 821 | 0.01052632 | 0.001191246 | 3.143458 | UUUUUU | UUUUUU |
| EIF4A3 | 1 | 821 | 0.01052632 | 0.001191246 | 3.143458 | UUUUUU | UUUUUU |
| U2AF2 | 2 | 1477 | 0.01578947 | 0.002141923 | 2.881984 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RALY | 2 | 1553 | 0.01578947 | 0.002252063 | 2.809644 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| CPEB2 | 3 | 2261 | 0.02105263 | 0.003278099 | 2.683069 | CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPCL1 | 2 | 2062 | 0.01578947 | 0.002989708 | 2.400887 | CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| YBX2 | 1 | 1480 | 0.01052632 | 0.002146271 | 2.294096 | ACAUCG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NONO | 1 | 1498 | 0.01052632 | 0.002172357 | 2.276668 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ENOX1 | 3 | 3195 | 0.02105263 | 0.004631656 | 2.184401 | AAGACA,CGGACA,GGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| SYNCRIP | 1 | 1634 | 0.01052632 | 0.002369448 | 2.151377 | UUUUUU | AAAAAA,UUUUUU |
| HNRNPC | 3 | 3471 | 0.02105263 | 0.005031636 | 2.064901 | CUUUUU,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZNF638 | 1 | 1773 | 0.01052632 | 0.002570888 | 2.033662 | GGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| MSI1 | 2 | 2770 | 0.01578947 | 0.004015744 | 1.975224 | AGGAGG,UAGGAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| KHDRBS2 | 1 | 1858 | 0.01052632 | 0.002694070 | 1.966141 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| PUM2 | 1 | 1890 | 0.01052632 | 0.002740445 | 1.941519 | UACAUC | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPU | 1 | 1965 | 0.01052632 | 0.002849135 | 1.885405 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPL | 4 | 5085 | 0.02631579 | 0.007370651 | 1.836065 | AAACAC,AACACA,ACACAA,CACAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| HNRNPA3 | 1 | 2140 | 0.01052632 | 0.003102746 | 1.762383 | AGGAGC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM46 | 3 | 4554 | 0.02105263 | 0.006601124 | 1.673217 | AAUGAA,AUGAAG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| TRA2A | 5 | 6871 | 0.03157895 | 0.009958930 | 1.664900 | AAAGAA,AAGAGG,GAAAGA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| HNRNPLL | 2 | 3534 | 0.01578947 | 0.005122936 | 1.623920 | ACUGCA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| PABPC5 | 1 | 2400 | 0.01052632 | 0.003479539 | 1.597033 | GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP3 | 3 | 4815 | 0.02105263 | 0.006979366 | 1.592833 | AAAAUC,AAACAC,AACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| FUS | 2 | 3648 | 0.01578947 | 0.005288145 | 1.578129 | UGGUGA,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| TRA2B | 5 | 7329 | 0.03157895 | 0.010622665 | 1.571817 | AAAGAA,AAGAAC,AGGAAA,GAAAGA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| TIA1 | 6 | 9206 | 0.03684211 | 0.013342821 | 1.465292 | CUCCUU,CUUUUU,UCCUUU,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SAMD4A | 2 | 3992 | 0.01578947 | 0.005786671 | 1.448157 | CGGGAA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| IGF2BP2 | 5 | 8408 | 0.03157895 | 0.012186356 | 1.373696 | AAAAUC,AAACAC,AACACA,GAACUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| QKI | 1 | 2805 | 0.01052632 | 0.004066466 | 1.372153 | UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPD | 1 | 2837 | 0.01052632 | 0.004112841 | 1.355793 | UUAGGA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ELAVL3 | 1 | 2867 | 0.01052632 | 0.004156317 | 1.340623 | UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| DAZAP1 | 3 | 5964 | 0.02105263 | 0.008644502 | 1.284146 | AGGAAA,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| SRSF10 | 8 | 13860 | 0.04736842 | 0.020087416 | 1.237634 | AAAGAA,AAAGAC,AAGACA,AAGAGG,ACAAAG,CAAAGA,GAAAGA,GAGGAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| ZRANB2 | 1 | 3173 | 0.01052632 | 0.004599773 | 1.194366 | GAGGUU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| KHDRBS3 | 1 | 3429 | 0.01052632 | 0.004970770 | 1.082459 | UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| FXR2 | 1 | 3434 | 0.01052632 | 0.004978016 | 1.080358 | GGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPDL | 4 | 8759 | 0.02631579 | 0.012695027 | 1.051665 | ACAGCA,ACUGCA,GGACUA,UCUGAC | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
RBP on BSJ (Exon Only)
Plot
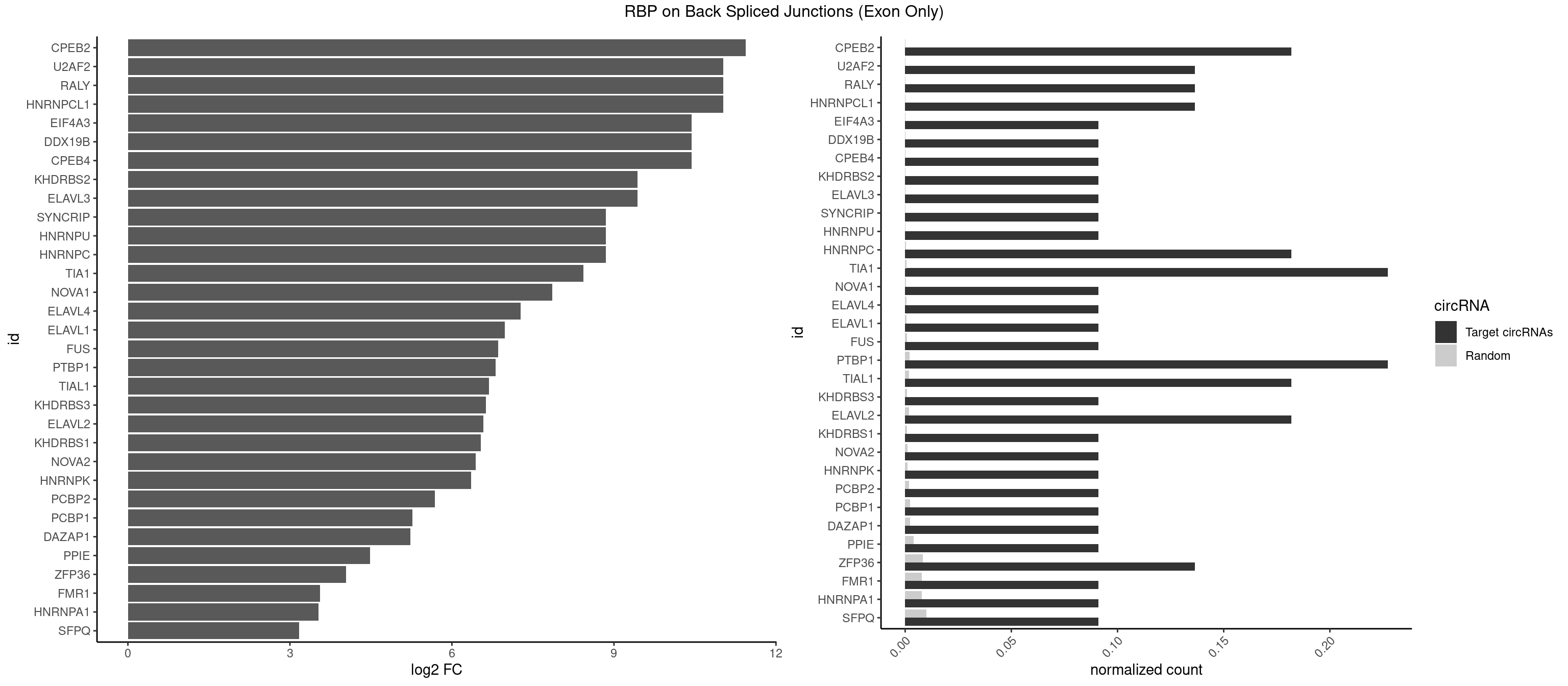
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CPEB2 | 3 | 0 | 0.18181818 | 6.549646e-05 | 11.438792 | CCUUUU,CUUUUU,UUUUUU | |
| HNRNPCL1 | 2 | 0 | 0.13636364 | 6.549646e-05 | 11.023754 | CUUUUU,UUUUUU | |
| RALY | 2 | 0 | 0.13636364 | 6.549646e-05 | 11.023754 | UUUUUC,UUUUUU | |
| U2AF2 | 2 | 0 | 0.13636364 | 6.549646e-05 | 11.023754 | UUUUUC,UUUUUU | |
| CPEB4 | 1 | 0 | 0.09090909 | 6.549646e-05 | 10.438792 | UUUUUU | |
| DDX19B | 1 | 0 | 0.09090909 | 6.549646e-05 | 10.438792 | UUUUUU | |
| EIF4A3 | 1 | 0 | 0.09090909 | 6.549646e-05 | 10.438792 | UUUUUU | |
| ELAVL3 | 1 | 1 | 0.09090909 | 1.309929e-04 | 9.438792 | UUUUUU | UAUUUU |
| KHDRBS2 | 1 | 1 | 0.09090909 | 1.309929e-04 | 9.438792 | UUUUUU | GAUAAA |
| HNRNPC | 3 | 5 | 0.18181818 | 3.929788e-04 | 8.853829 | CUUUUU,UUUUUC,UUUUUU | GGAUAU,GGAUUC,GGGUAC |
| HNRNPU | 1 | 2 | 0.09090909 | 1.964894e-04 | 8.853829 | UUUUUU | AAAAAA |
| SYNCRIP | 1 | 2 | 0.09090909 | 1.964894e-04 | 8.853829 | UUUUUU | AAAAAA |
| TIA1 | 4 | 9 | 0.22727273 | 6.549646e-04 | 8.438792 | CUUUUU,UCCUUU,UUUUUC,UUUUUU | AUUUUG,GUUUUG,GUUUUU,UAUUUU,UUCUCC,UUUUGG,UUUUGU |
| NOVA1 | 1 | 5 | 0.09090909 | 3.929788e-04 | 7.853829 | UUUUUU | ACCACC,CAGUCA,CAUUCA,UCAGUC |
| ELAVL4 | 1 | 8 | 0.09090909 | 5.894682e-04 | 7.268867 | UUUUUU | AAAAAA,AUCUAA,UAUUUU,UCUAAU |
| ELAVL1 | 1 | 10 | 0.09090909 | 7.204611e-04 | 6.979360 | UUUUUU | UAUUUU,UGGUUU,UUAGUU,UUGGUU,UUUAGU,UUUGGU,UUUGUU |
| FUS | 1 | 11 | 0.09090909 | 7.859576e-04 | 6.853829 | UUUUUU | AAAAAA,CGGUGG,GGGUGA,GGGUGG,GGGUGU |
| PTBP1 | 4 | 30 | 0.22727273 | 2.030390e-03 | 6.806524 | CCUUUU,CUUUUU,UUUUUC,UUUUUU | AGCUGU,CAUCUU,CCAUCU,CCUCUU,CUAUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUUCU,GGCUCC,GUCUUU,UACUUU,UAUUUU,UCCUCU,UCUAUC,UCUCUU,UCUUCU,UCUUUC,UUACUU,UUCUUC,UUCUUG,UUUCUU |
| TIAL1 | 3 | 26 | 0.18181818 | 1.768405e-03 | 6.683904 | CUUUUU,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUG,CAGUUU,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUCAGU,UUUCAG |
| KHDRBS3 | 1 | 13 | 0.09090909 | 9.169505e-04 | 6.631437 | UUUUUU | AAAUAA,AGAUAA,AUAAAG,AUUAAA,GAUAAA,UAAAUA,UUAAAC |
| ELAVL2 | 3 | 28 | 0.18181818 | 1.899397e-03 | 6.580811 | CUUUUU,UUUUUC,UUUUUU | AAUUUA,AAUUUU,AUAUUU,AUUUAA,AUUUUG,CUUUCU,GAUUUU,GUAUUG,GUUUUU,UACUUU,UAGUUA,UAUAUA,UAUAUU,UAUGUU,UAUUGA,UAUUUU,UGUAUU,UUAAGU,UUACUU,UUAGUU,UUAUUG,UUUACU,UUUAGU,UUUCUU,UUUUAG |
| KHDRBS1 | 1 | 14 | 0.09090909 | 9.824469e-04 | 6.531901 | UUUUUU | AUUUAA,CUAAAA,GAAAAC,UAAAAA,UAAAAG,UUAAAA,UUAAAU |
| NOVA2 | 1 | 15 | 0.09090909 | 1.047943e-03 | 6.438792 | UUUUUU | ACCACC,AGACAU,AGAUCA,AGGCAU,AGUCAU,CUAGAU,GAGACA,GAGGCA,GAGUCA,UAGAUC |
| HNRNPK | 1 | 16 | 0.09090909 | 1.113440e-03 | 6.351329 | UUUUUU | AAAAAA,ACCCCA,ACCCCC,ACCCGA,CAAACC,CAUACC,CCAACC,CCAUAC,CCCCAC,CCCCCU,CCCCUU,GCCCAC,GCCCAG,UCCCAC |
| PCBP2 | 1 | 26 | 0.09090909 | 1.768405e-03 | 5.683904 | UUUUUU | AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,AUUAAA,CCAUUC,CCCCCU,CCCUCC,CCCUUA,CCUCCC,CUCCCU,CUUCCA,UAACCC,UUAGAG,UUAGGA,UUAGGG |
| PCBP1 | 1 | 35 | 0.09090909 | 2.357873e-03 | 5.268867 | UUUUUU | AAAAAA,AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,ACCCUC,AUUAAA,CAAACC,CAUACC,CCAACC,CCACCC,CCAUAC,CCAUUC,CCCACC,CCCCAC,CCCUCC,CCCUUA,CCUCCC,CCUCUU,UUAGAG,UUAGGA,UUAGGG |
| DAZAP1 | 1 | 36 | 0.09090909 | 2.423369e-03 | 5.229338 | UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| PPIE | 1 | 61 | 0.09090909 | 4.060781e-03 | 4.484596 | UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAU,AUAAUA,AUAUAA,AUAUAU,AUAUUU,AUUAAA,AUUUAA,UAAAAA,UAAAUA,UAAAUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUU,UUAAAA,UUAAAU,UUAUUA |
| ZFP36 | 2 | 126 | 0.13636364 | 8.318051e-03 | 4.035070 | CCUUUU,UCCUUU | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
| FMR1 | 1 | 117 | 0.09090909 | 7.728583e-03 | 3.556149 | UUUUUU | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| HNRNPA1 | 1 | 119 | 0.09090909 | 7.859576e-03 | 3.531901 | UUUUUU | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
| SFPQ | 1 | 153 | 0.09090909 | 1.008646e-02 | 3.172005 | UUUUUU | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
RBP on BSJ (Exon and Intron)
Plot
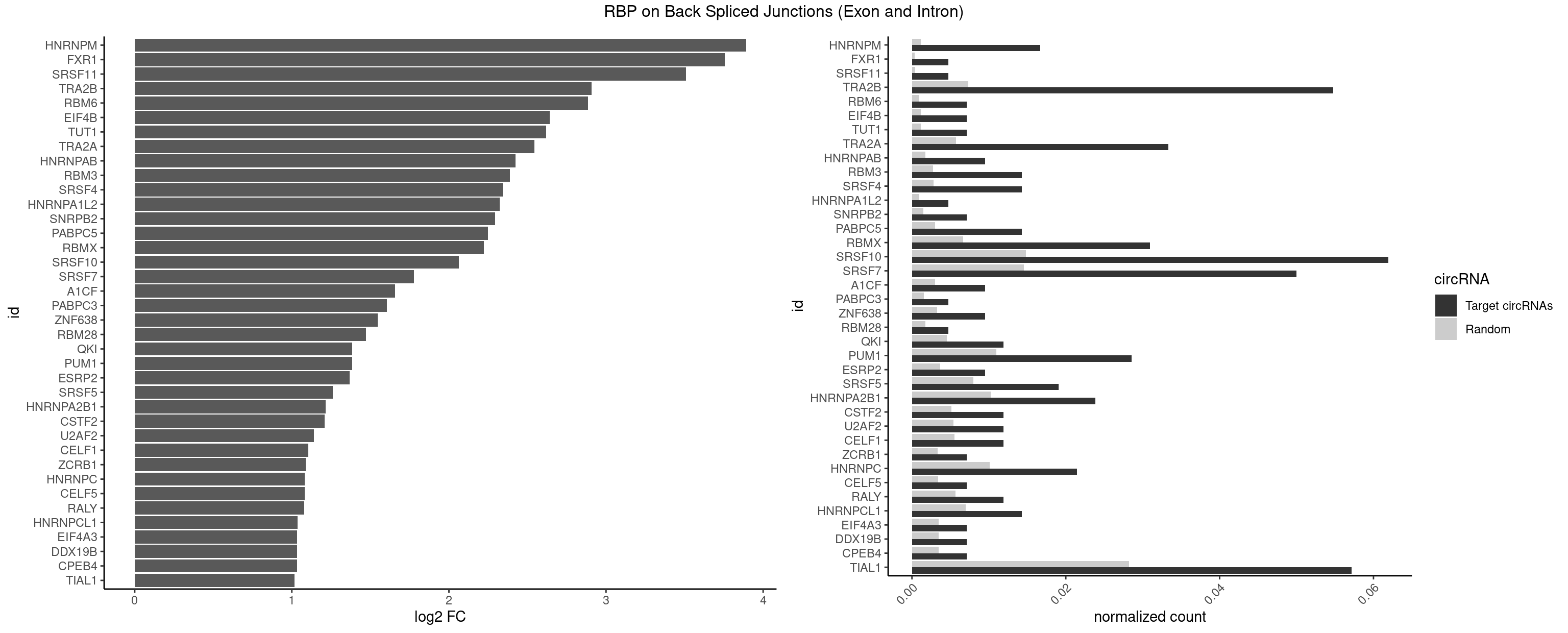
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPM | 6 | 222 | 0.016666667 | 0.0011235389 | 3.890844 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| TRA2B | 22 | 1447 | 0.054761905 | 0.0072954454 | 2.908105 | AAAGAA,AAGAAG,AAGAAU,AAGGAA,AGAAGG,AGGAAA,AGGAAG,GAAAGA,GAAGAA,GAAGGA,GGAAGG,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| TRA2A | 13 | 1133 | 0.033333333 | 0.0057134220 | 2.544539 | AAAGAA,AAGAAA,AAGAAG,AGAAAG,AGGAAG,GAAAGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| HNRNPAB | 3 | 351 | 0.009523810 | 0.0017734784 | 2.424957 | ACAAAG,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 5 | 541 | 0.014285714 | 0.0027307537 | 2.387202 | AAAACU,AAUACU,AGACUA,GAAACG,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SRSF4 | 5 | 558 | 0.014285714 | 0.0028164047 | 2.342647 | AAGAAG,AGGAAG,GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | GUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PABPC5 | 5 | 596 | 0.014285714 | 0.0030078597 | 2.247764 | AGAAAG,AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBMX | 12 | 1316 | 0.030952381 | 0.0066354293 | 2.221789 | AAGAAG,AAGGAA,AGAAGG,AGGAAG,GAAGGA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SRSF10 | 25 | 2937 | 0.061904762 | 0.0148024990 | 2.064210 | AAAGAA,AAAGAG,AAGAAA,AAGAAG,AAGAGA,AAGGAA,ACAAAG,AGAGAA,AGAGAC,AGAGAG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SRSF7 | 20 | 2893 | 0.050000000 | 0.0145808142 | 1.777857 | AAGAAG,AAUGAU,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGGAAG,AUUGAU,GAAGAA,GACAAA,GACUAC,GAGACU,GAGAGA,GGAAGG,GGACAA,UGGACA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | AUAAUU,CAGUAU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACACUA,CUAACA,UCAUAU,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PUM1 | 11 | 2172 | 0.028571429 | 0.0109482064 | 1.383879 | AAUGUU,AAUUGU,AUUGUA,CUUGUA,UAAUGU,UACAUA,UGUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SRSF5 | 7 | 1578 | 0.019047619 | 0.0079554615 | 1.259593 | AAGAAG,AGGAAG,AUAAAG,GAAGAA,GGAAGA,UACAUA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| HNRNPA2B1 | 9 | 2033 | 0.023809524 | 0.0102478839 | 1.216213 | AAGAAG,AAGGAA,AGACUA,GAAGGA,GUAGGG | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUUU,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | GUUUGU,UGUGUU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPC | 8 | 2006 | 0.021428571 | 0.0101118501 | 1.083489 | AUUUUU,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.0069629182 | 1.036809 | AUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| TIAL1 | 23 | 5597 | 0.057142857 | 0.0282043531 | 1.018655 | AAAUUU,AAUUUU,AGUUUU,AUUUUU,CUUUUA,GUUUUC,UAAAUU,UAUUUU,UUAAAU,UUUAUU,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.