circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000140470:-:15:100254135:100281401
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000140470:-:15:100254135:100281401 | ENSG00000140470 | ENST00000268070 | - | 15 | 100254136 | 100281401 | 459 | AAAAGAAGAAGCCGACGUGGGGCAGGCCUUCGCGGGACUGGCGGGAGCGGAGGAACGCUAUCCGGCUCACCAGCGAGCACACGGUGGAGACCCUGGUGGUGGCCGACGCCGACAUGGUGCAGUACCACGGGGCCGAGGCCGCCCAGAGGUUCAUCCUGACCGUCAUGAACAUGGUAUACAAUAUGUUUCAGCACCAGAGCCUGGGGAUUAAAAUUAACAUUCAAGUGACCAAGCUUGUCCUGCUACGACAACGUCCCGCUAAGUUGUCCAUUGGGCACCAUGGUGAGCGGUCCCUGGAGAGCUUCUGUCACUGGCAGAACGAGGAGUAUGGAGGAGCGCGAUACCUCGGCAAUAACCAGGUUCCCGGCGGGAAGGACGACCCGCCCCUGGUGGAUGCUGCCGUGUUUGUGACCAGGACAGAUUUCUGUGUACACAAGGAUGAACCGUGUGACACUGUUGAAAAGAAGAAGCCGACGUGGGGCAGGCCUUCGCGGGACUGGCGGGAGCGG | circ |
| ENSG00000140470:-:15:100254135:100281401 | ENSG00000140470 | ENST00000268070 | - | 15 | 100254136 | 100281401 | 22 | UGACACUGUUGAAAAGAAGAAG | bsj |
| ENSG00000140470:-:15:100254135:100281401 | ENSG00000140470 | ENST00000268070 | - | 15 | 100281392 | 100281601 | 210 | CCCUUCUUUUGGGCCAGCACCCCCCUACGUGAGUGAACUCGGGGCACCCACAUGACGGUGGAGAUUGCUGGGUGCUGGGCCCCUCUAGGCCUGUCGAGACUGACCACUCUCUCGGGCAGGCUUGACAGAAGGCCUGUUCACUGCAUGGUUUUGGAAGUCAGUAAGCCAAGGACCGCACAAAUGUUUCCAUCAUUUUCUAGAAAAGAAGAA | ie_up |
| ENSG00000140470:-:15:100254135:100281401 | ENSG00000140470 | ENST00000268070 | - | 15 | 100253936 | 100254145 | 210 | GACACUGUUGGUAAGUUUAGAGUUGAAAUAAUAAUCUAAGGGGCUGAUACACCCUGGGAGAAUCUGGAGACUUCCUGGUCACAGUGGAGGAGCUGUCCUCAAACAAGCACUGCACAUUCUGUUGCCUUCCUUCCUGUAAGUUGGUCUUUGACUUUCCUUUUUUUAUUUUCCUGAAAUGAUUUGAAGUUAUUGAUGGCAGAAGGCAUCUAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
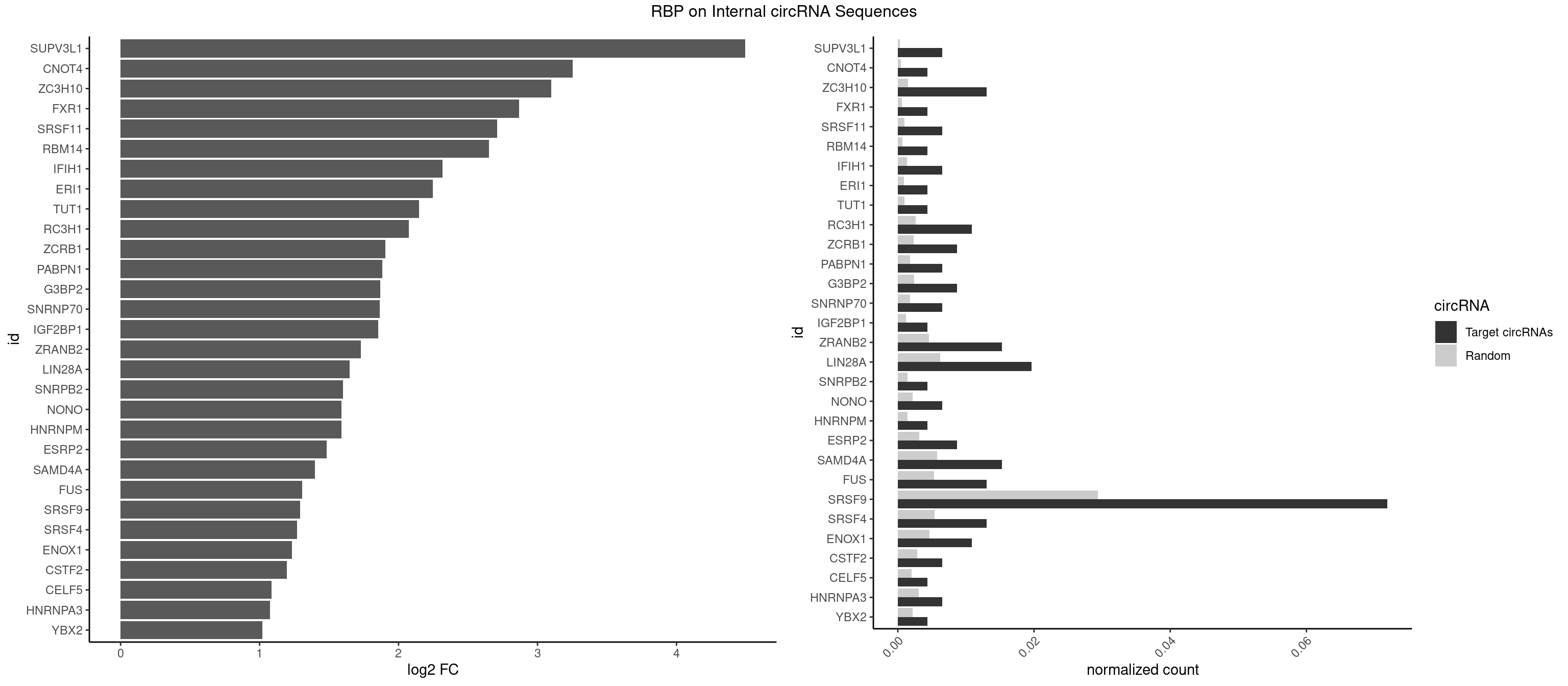
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 2 | 199 | 0.006535948 | 0.0002898408 | 4.495064 | CCGCCC | CCGCCC |
| CNOT4 | 1 | 314 | 0.004357298 | 0.0004564992 | 3.254750 | GACAGA | GACAGA |
| ZC3H10 | 5 | 1053 | 0.013071895 | 0.0015274610 | 3.097261 | CAGCGA,CCAGCG,GAGCGC,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| FXR1 | 1 | 411 | 0.004357298 | 0.0005970720 | 2.867457 | ACGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRSF11 | 2 | 688 | 0.006535948 | 0.0009985015 | 2.710560 | AAGAAG | AAGAAG |
| RBM14 | 1 | 478 | 0.004357298 | 0.0006941687 | 2.650076 | CGCGGG | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| IFIH1 | 2 | 904 | 0.006535948 | 0.0013115296 | 2.317146 | GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ERI1 | 1 | 632 | 0.004357298 | 0.0009173461 | 2.247896 | UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 1 | 678 | 0.004357298 | 0.0009840095 | 2.146690 | CGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RC3H1 | 4 | 1786 | 0.010893246 | 0.0025897275 | 2.072562 | CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZCRB1 | 3 | 1605 | 0.008714597 | 0.0023274215 | 1.904701 | AAUUAA,GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PABPN1 | 2 | 1222 | 0.006535948 | 0.0017723764 | 1.882711 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| G3BP2 | 3 | 1644 | 0.008714597 | 0.0023839405 | 1.870086 | AGGAUG,GGAUGA,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SNRNP70 | 2 | 1237 | 0.006535948 | 0.0017941145 | 1.865124 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| IGF2BP1 | 1 | 831 | 0.004357298 | 0.0012057377 | 1.853518 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ZRANB2 | 6 | 3173 | 0.015250545 | 0.0045997733 | 1.729226 | GAGGUU,GGUGGU,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| LIN28A | 8 | 4315 | 0.019607843 | 0.0062547643 | 1.648403 | AGGAGU,CGGAGG,GGAGGA,GGAGUA,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SNRPB2 | 1 | 991 | 0.004357298 | 0.0014376103 | 1.599761 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| NONO | 2 | 1498 | 0.006535948 | 0.0021723567 | 1.589135 | AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| HNRNPM | 1 | 999 | 0.004357298 | 0.0014492040 | 1.588173 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ESRP2 | 3 | 2150 | 0.008714597 | 0.0031172377 | 1.483166 | GGGAAG,GGGGAU,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SAMD4A | 6 | 3992 | 0.015250545 | 0.0057866714 | 1.398055 | CGGGAA,CGGGAC,CUGGCA,GCGGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| FUS | 5 | 3648 | 0.013071895 | 0.0052881452 | 1.305635 | CGGUGG,UGGUGA,UGGUGG | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| SRSF9 | 32 | 20254 | 0.071895425 | 0.0293536261 | 1.292361 | AGGAAC,AGGACA,AGGAGC,AUGAAC,GAAGCC,GAAGGA,GGAACG,GGAAGG,GGACAG,GGAGAG,GGAGCG,GGAGGA,GGAUGC,GGGAGC,GGUGCA,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGACCA,UGACCG,UGAGCG,UGGAGG,UGGUGC,UGGUGG | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| SRSF4 | 5 | 3740 | 0.013071895 | 0.0054214720 | 1.269712 | AAGAAG,AGAAGA,GAAGAA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ENOX1 | 4 | 3195 | 0.010893246 | 0.0046316558 | 1.233834 | AGGACA,GGACAG,UAUACA,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| CSTF2 | 2 | 1967 | 0.006535948 | 0.0028520334 | 1.196406 | GUGUUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF5 | 1 | 1415 | 0.004357298 | 0.0020520728 | 1.086352 | GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPA3 | 2 | 2140 | 0.006535948 | 0.0031027457 | 1.074851 | AGGAGC,CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| YBX2 | 1 | 1480 | 0.004357298 | 0.0021462711 | 1.021602 | ACAACG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
RBP on BSJ (Exon Only)
Plot
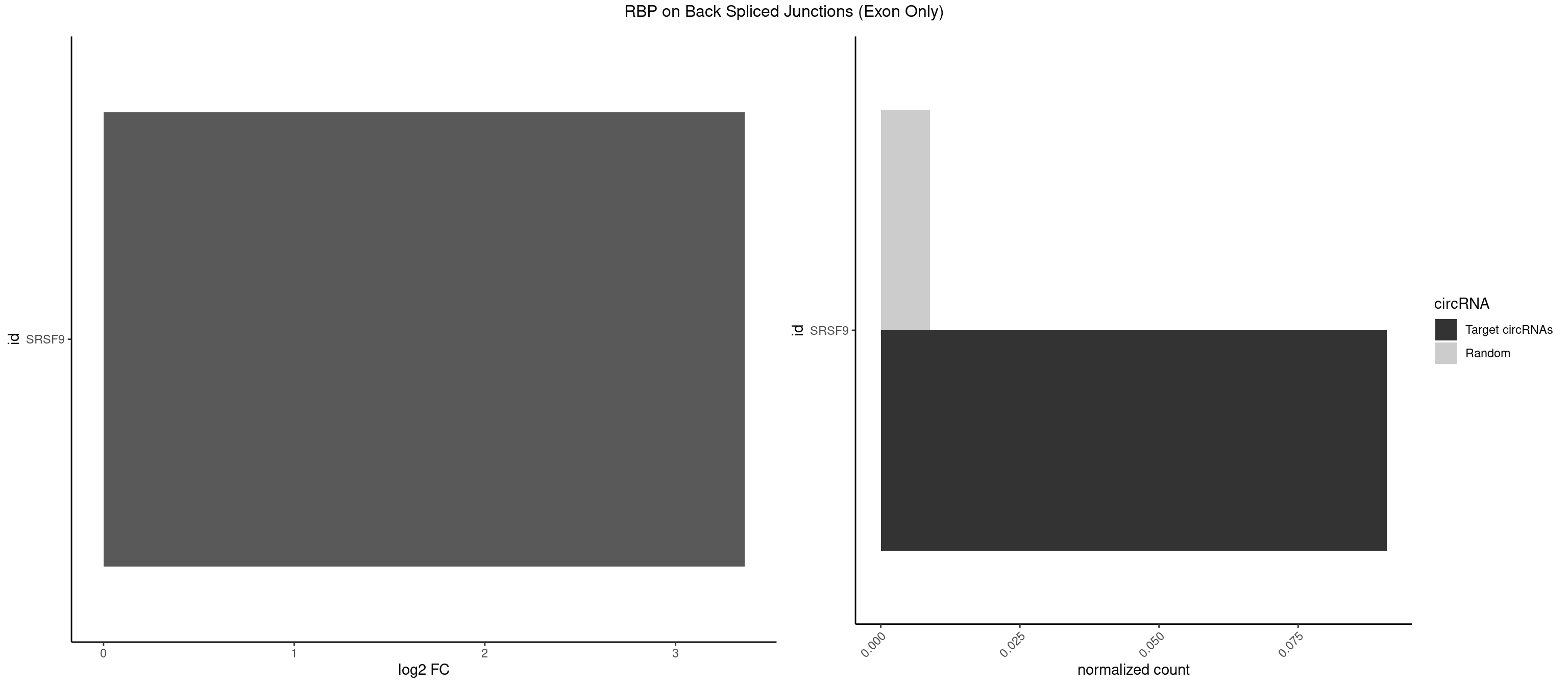
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF9 | 1 | 134 | 0.09090909 | 0.008842023 | 3.361976 | UGAAAA | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
RBP on BSJ (Exon and Intron)
Plot
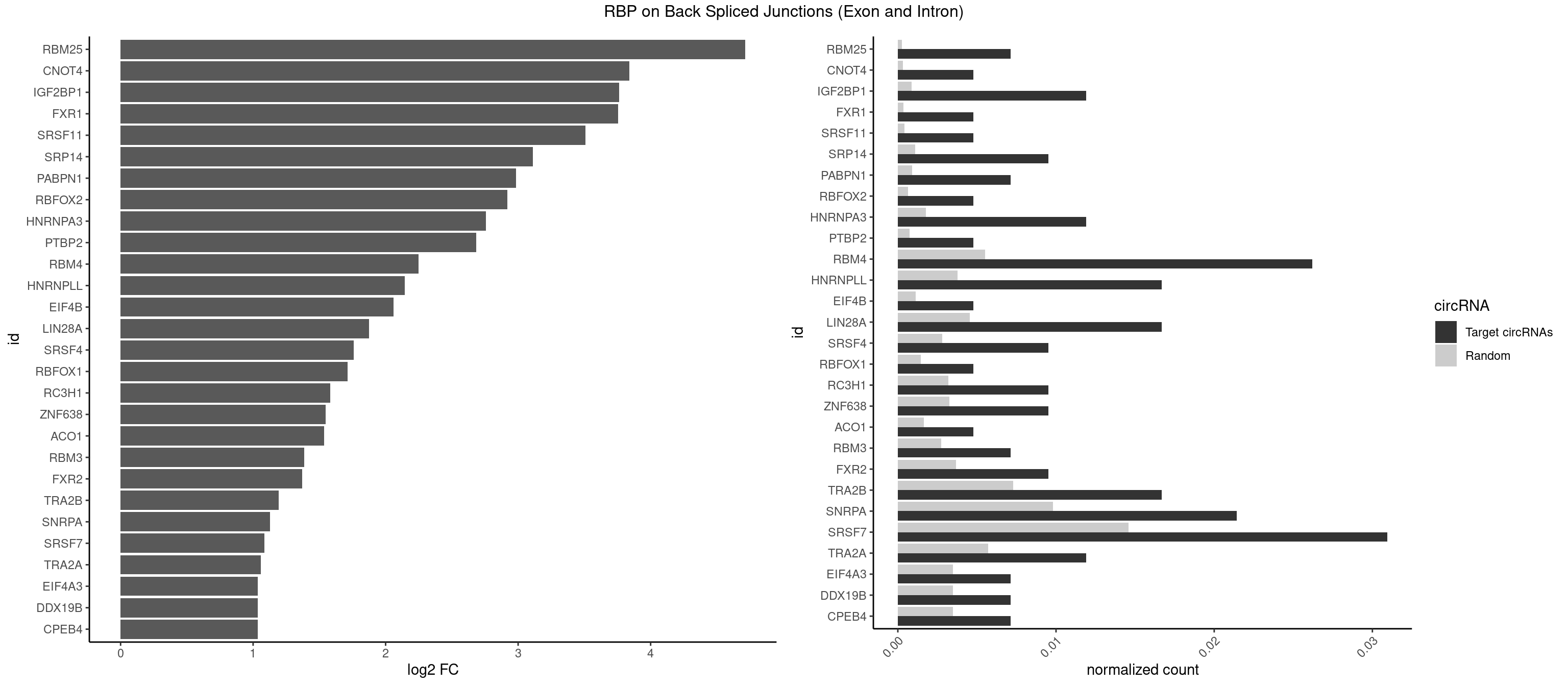
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 2 | 53 | 0.007142857 | 0.0002720677 | 4.714464 | CGGGCA,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| IGF2BP1 | 4 | 173 | 0.011904762 | 0.0008766626 | 3.763373 | AAGCAC,AGCACC,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| SRP14 | 3 | 218 | 0.009523810 | 0.0011033857 | 3.109602 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| HNRNPA3 | 4 | 349 | 0.011904762 | 0.0017634019 | 2.755106 | AGGAGC,CAAGGA,CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| RBM4 | 10 | 1094 | 0.026190476 | 0.0055169287 | 2.247105 | CCUUCC,CCUUCU,CUUCCU,CUUCUU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| HNRNPLL | 6 | 748 | 0.016666667 | 0.0037736800 | 2.142922 | ACCGCA,ACUGCA,CAAACA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| LIN28A | 6 | 900 | 0.016666667 | 0.0045395002 | 1.876360 | GGAGAA,GGAGAU,GGAGGA,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SRSF4 | 3 | 558 | 0.009523810 | 0.0028164047 | 1.757684 | AAGAAG,AGAAGA,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CCCUUC,CCUUCU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| FXR2 | 3 | 730 | 0.009523810 | 0.0036829907 | 1.370661 | GACAGA,UGACAG,UGACGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| TRA2B | 6 | 1447 | 0.016666667 | 0.0072954454 | 1.191898 | AAAGAA,AAGAAG,AGAAGA,AGAAGG,GAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SNRPA | 8 | 1947 | 0.021428571 | 0.0098145909 | 1.126536 | GGAGAU,GUUUCC,UGCACA,UUCCUG,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| SRSF7 | 12 | 2893 | 0.030952381 | 0.0145808142 | 1.085979 | AAGAAG,AAGGAC,AAUGAU,AGAAGA,AUUGAU,CGAGAC,GAAGAA,GAAGGC,GAGACU,UCGAGA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AAAGAA,AAGAAG,AGAAGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.