circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000198722:+:9:35228014:35259050
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198722:+:9:35228014:35259050 | ENSG00000198722 | ENST00000635942 | + | 9 | 35228015 | 35259050 | 504 | UUAAAAGGGCCAAAUUCCAGGGUUCACCAGAUAAAUUUAACACAUAUGUGACCCUGAAAGUACAGAAUGUGAAGAGCACAACUGUAGCAGUUCGUGGUGAUCAGCCUUCCUGGGAACAGGAUUUCAUGUUUGAGAUUAGUCGCCUGGACCUGGGUCUAAGUGUGGAGGUAUGGAACAAAGGACUGAUCUGGGACACCAUGGUGGGGACUGUGUGGAUUGCGCUGAAGACUAUUCGUCAGUCGGAUGAGGAAGGGCCUGGGGAAUGGUCCACAUUAGAGGCAGAGACGUUAAUGAAAGACGAUGAGAUCUGUGGAACUAGAAACCCAACUCCUCAUAAAAUUUUGCUUGAUACAAGAUUUGAGUUGCCUUUUGAUAUCCCAGAGGAGGAAGCCAGAUAUUGGACCUACAAAUGGGAGCAAAUCAAUGCCUUGGGAGCUGACAAUGAGUAUUCUAGUCAAGAAGAAAGCCAGAGGAAGCCAUUGCCCACUGCUGCCGCCCAGUGUUUUAAAAGGGCCAAAUUCCAGGGUUCACCAGAUAAAUUUAACACAUAUGUG | circ |
| ENSG00000198722:+:9:35228014:35259050 | ENSG00000198722 | ENST00000635942 | + | 9 | 35228015 | 35259050 | 22 | CGCCCAGUGUUUUAAAAGGGCC | bsj |
| ENSG00000198722:+:9:35228014:35259050 | ENSG00000198722 | ENST00000635942 | + | 9 | 35227815 | 35228024 | 210 | UGAGAUCAGAUUGACUUUUAAGGUCUCGUCUGACUCUUUGCCUUUCAAGUUUUGAUCAGAAGUCUCAGCUUUUUUUCUCCAGGUUUAGAUUUAACAAUCUAAACCUAGUUCUAACAUUGGUAUGUAGUGCCUAAUAAAUGUUGCUUAAGUAAAAUGAACUUGGAAACAUUCUUCAUGGUAUCCUCUUUUUUCUCUUGCAGUUAAAAGGGC | ie_up |
| ENSG00000198722:+:9:35228014:35259050 | ENSG00000198722 | ENST00000635942 | + | 9 | 35259041 | 35259250 | 210 | GCCCAGUGUUGUAAGUGAGAAACUUGUCUAUGAAUCUCUUUUAAUUGUAGCUUUCUGUCGCUUCUCUGAACAUGGGUUAUUCUGAGCUGGGGAGGGUGGAGAUGUCAUGUGUGUAAGCAGAAGAUAUUCCUGAAGCACAUCUGUUCAGGCGCCUUUCUCUGAUGGCUGUCUAUCUAUUAGCUUUAGGGAGACGUUUUUCGGUUUUGUAUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
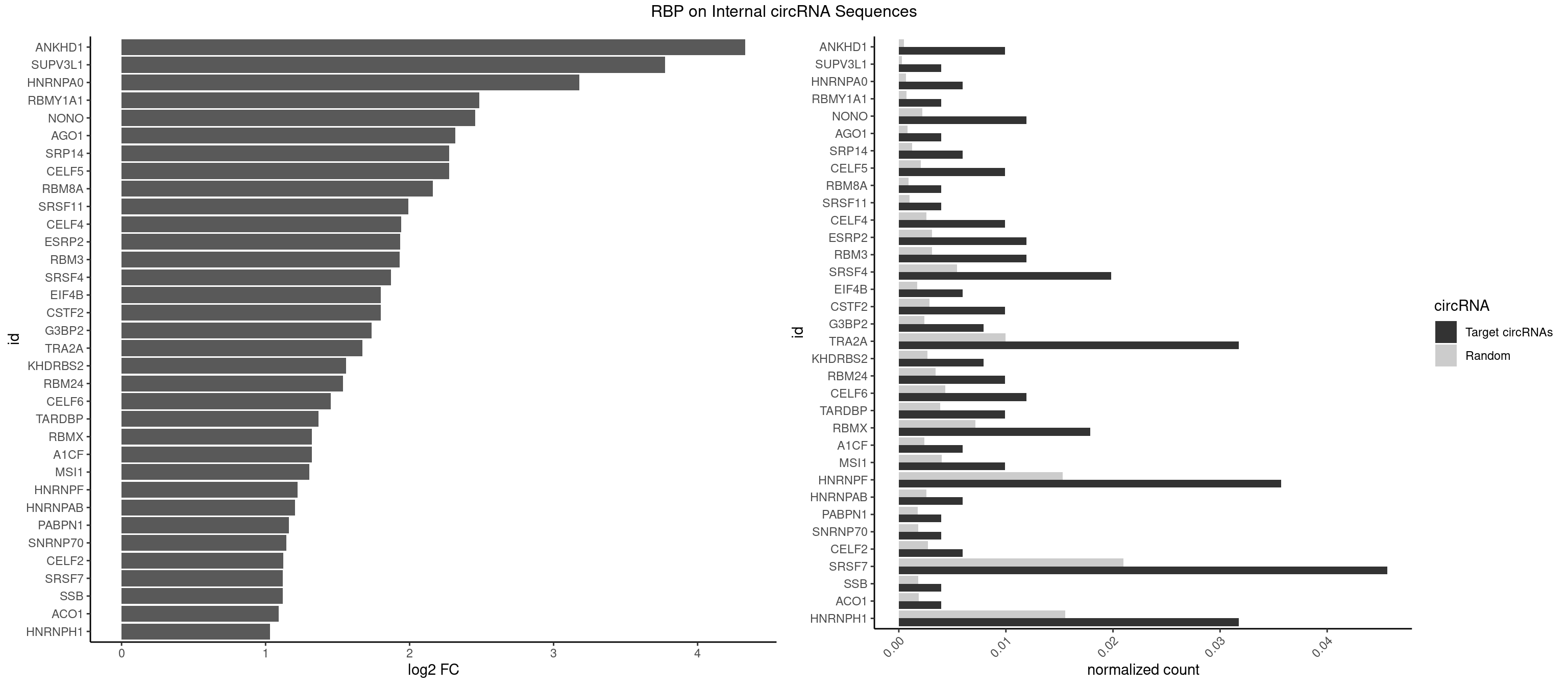
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 4 | 339 | 0.009920635 | 0.0004927293 | 4.331565 | AGACGA,AGACGU,GACGAU,GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SUPV3L1 | 1 | 199 | 0.003968254 | 0.0002898408 | 3.775172 | CCGCCC | CCGCCC |
| HNRNPA0 | 2 | 453 | 0.005952381 | 0.0006579386 | 3.177442 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMY1A1 | 1 | 489 | 0.003968254 | 0.0007101099 | 2.482390 | ACAAGA | ACAAGA,CAAGAC |
| NONO | 5 | 1498 | 0.011904762 | 0.0021723567 | 2.454206 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| AGO1 | 1 | 548 | 0.003968254 | 0.0007956130 | 2.318366 | GAGGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SRP14 | 2 | 847 | 0.005952381 | 0.0012289250 | 2.276070 | CGCCUG,CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| CELF5 | 4 | 1415 | 0.009920635 | 0.0020520728 | 2.273351 | GUGUGG,GUGUUU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM8A | 1 | 611 | 0.003968254 | 0.0008869128 | 2.161640 | UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| SRSF11 | 1 | 688 | 0.003968254 | 0.0009985015 | 1.990668 | AAGAAG | AAGAAG |
| CELF4 | 4 | 1782 | 0.009920635 | 0.0025839306 | 1.940865 | GUGUGG,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ESRP2 | 5 | 2150 | 0.011904762 | 0.0031172377 | 1.933199 | GGGAAU,GGGGAA,UGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM3 | 5 | 2152 | 0.011904762 | 0.0031201361 | 1.931858 | AAGACG,AAGACU,AGACGA,AGACUA,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SRSF4 | 9 | 3740 | 0.019841270 | 0.0054214720 | 1.871748 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| EIF4B | 2 | 1179 | 0.005952381 | 0.0017100607 | 1.799419 | GUCGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| CSTF2 | 4 | 1967 | 0.009920635 | 0.0028520334 | 1.798442 | GUGUUU,UGUGUG,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| G3BP2 | 3 | 1644 | 0.007936508 | 0.0023839405 | 1.735156 | AGGAUU,GGAUGA,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| TRA2A | 15 | 6871 | 0.031746032 | 0.0099589296 | 1.672514 | AAGAAA,AAGAAG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| KHDRBS2 | 3 | 1858 | 0.007936508 | 0.0026940701 | 1.558717 | AUAAAA,GAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RBM24 | 4 | 2357 | 0.009920635 | 0.0034172229 | 1.537608 | AGUGUG,GUGUGG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF6 | 5 | 2997 | 0.011904762 | 0.0043447134 | 1.454206 | GUGGGG,GUGGUG,GUGUGG,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| TARDBP | 4 | 2654 | 0.009920635 | 0.0038476365 | 1.366460 | GAAUGG,GAAUGU,UGUGUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBMX | 8 | 4925 | 0.017857143 | 0.0071387787 | 1.322752 | AAGAAG,AAGUGU,AGGAAG,AGUGUU,AUCCCA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| A1CF | 2 | 1642 | 0.005952381 | 0.0023810421 | 1.321874 | GAUCAG,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| MSI1 | 4 | 2770 | 0.009920635 | 0.0040157442 | 1.304765 | AGGAAG,AGGAGG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| HNRNPF | 17 | 10561 | 0.035714286 | 0.0153064921 | 1.222358 | AAUGUG,AGGAAG,AUGGGA,CUGGGG,GAAGGG,GAAUGU,GAGGAA,GGAAGG,GGAAUG,GGAGGA,GGGAAU,GGGGAA,UGGGAA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPAB | 2 | 1782 | 0.005952381 | 0.0025839306 | 1.203900 | AAAGAC,ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPN1 | 1 | 1222 | 0.003968254 | 0.0017723764 | 1.162819 | AGAAGA | AAAAGA,AGAAGA |
| SNRNP70 | 1 | 1237 | 0.003968254 | 0.0017941145 | 1.145232 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF2 | 2 | 1886 | 0.005952381 | 0.0027346479 | 1.122112 | UAUGUG,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SRSF7 | 22 | 14481 | 0.045634921 | 0.0209873716 | 1.120617 | AAAGGA,AAGAAG,AAGGAC,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAC,AGAGGA,AGAUCU,AGGAAG,CAGAGA,GAAGAA,GACGAU,GAGAUC,GAGGAA,GGAAGG | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| SSB | 1 | 1260 | 0.003968254 | 0.0018274462 | 1.118675 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ACO1 | 1 | 1283 | 0.003968254 | 0.0018607779 | 1.092599 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPH1 | 15 | 10717 | 0.031746032 | 0.0155325680 | 1.031280 | AAUGUG,AGGAAG,CUGGGG,GAAGGG,GAAUGU,GAGGAA,GGAAGG,GGAAUG,GGAGGA,GGGAAU,GGGGAA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
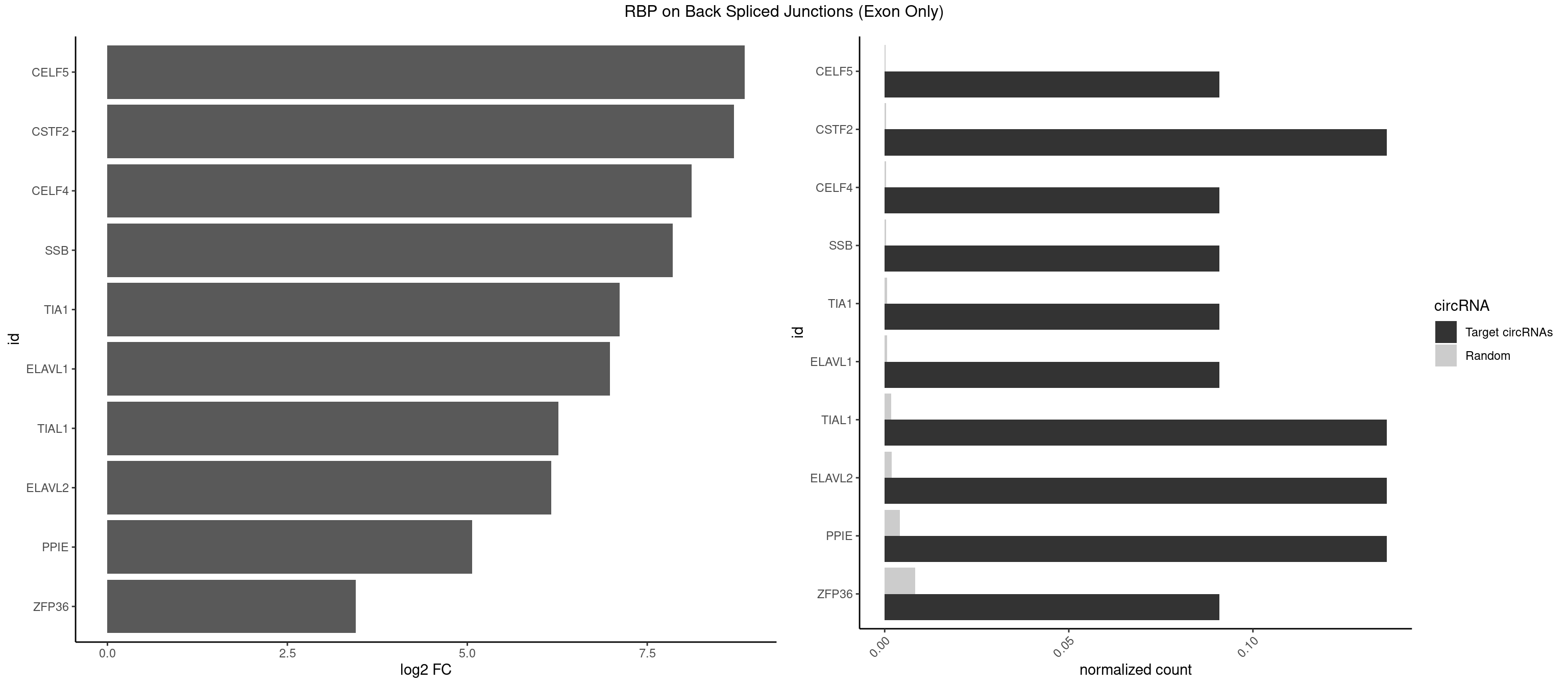
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CELF5 | 1 | 2 | 0.09090909 | 0.0001964894 | 8.853829 | GUGUUU | GUGUUG,UGUGUG |
| CSTF2 | 2 | 4 | 0.13636364 | 0.0003274823 | 8.701826 | GUGUUU,UGUUUU | GUGUUG,GUUUUG,UGUGUG,UGUUUG |
| CELF4 | 1 | 4 | 0.09090909 | 0.0003274823 | 8.116864 | GUGUUU | GGUGUG,GUGUUG,UGUGUG |
| SSB | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | UGUUUU | CUGUUU,GCUGUU,UGCUGU |
| TIA1 | 1 | 9 | 0.09090909 | 0.0006549646 | 7.116864 | GUUUUA | AUUUUG,GUUUUG,GUUUUU,UAUUUU,UUCUCC,UUUUGG,UUUUGU |
| ELAVL1 | 1 | 10 | 0.09090909 | 0.0007204611 | 6.979360 | UGUUUU | UAUUUU,UGGUUU,UUAGUU,UUGGUU,UUUAGU,UUUGGU,UUUGUU |
| TIAL1 | 2 | 26 | 0.13636364 | 0.0017684045 | 6.268867 | UUUAAA,UUUUAA | AAAUUU,AAUUUU,AGUUUU,AUUUUG,CAGUUU,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUCAGU,UUUCAG |
| ELAVL2 | 2 | 28 | 0.13636364 | 0.0018993974 | 6.165773 | GUUUUA,UUUUAA | AAUUUA,AAUUUU,AUAUUU,AUUUAA,AUUUUG,CUUUCU,GAUUUU,GUAUUG,GUUUUU,UACUUU,UAGUUA,UAUAUA,UAUAUU,UAUGUU,UAUUGA,UAUUUU,UGUAUU,UUAAGU,UUACUU,UUAGUU,UUAUUG,UUUACU,UUUAGU,UUUCUU,UUUUAG |
| PPIE | 2 | 61 | 0.13636364 | 0.0040607807 | 5.069558 | UUUAAA,UUUUAA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAU,AUAAUA,AUAUAA,AUAUAU,AUAUUU,AUUAAA,AUUUAA,UAAAAA,UAAAUA,UAAAUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUU,UUAAAA,UUAAAU,UUAUUA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.0083180508 | 3.450107 | UUUAAA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
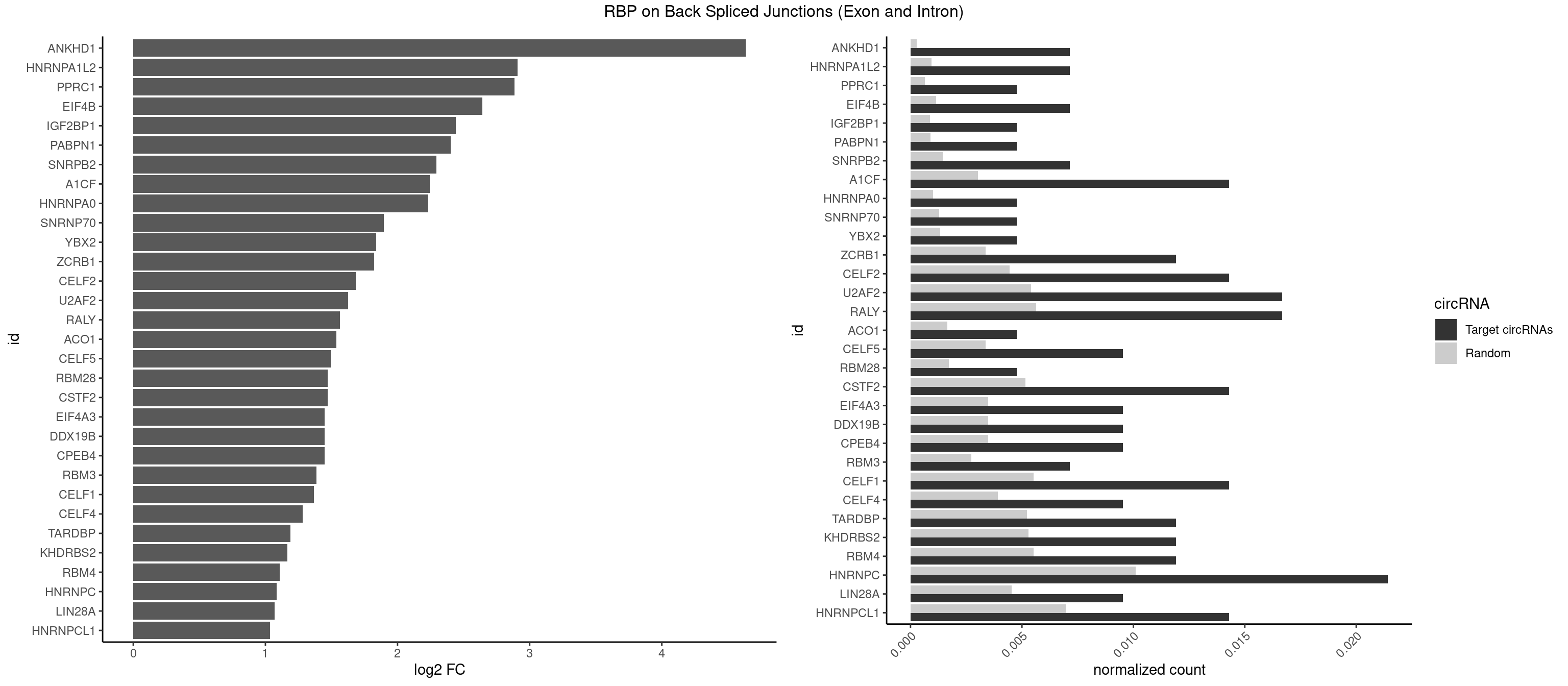
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 56 | 0.007142857 | 0.0002871826 | 4.636461 | AGACGU,GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| PPRC1 | 1 | 127 | 0.004761905 | 0.0006449012 | 2.884389 | GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| A1CF | 5 | 598 | 0.014285714 | 0.0030179363 | 2.242939 | GAUCAG,UAAUUG,UGAUCA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | AUUUAA,GAUUUA,GCUUAA,GGUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CELF2 | 5 | 881 | 0.014285714 | 0.0044437727 | 1.684716 | AUGUGU,GUAUGU,GUGUGU,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| U2AF2 | 6 | 1071 | 0.016666667 | 0.0054010480 | 1.625654 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RALY | 6 | 1117 | 0.016666667 | 0.0056328094 | 1.565039 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGU,GUGUUG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.0051541717 | 1.470761 | GUGUGU,GUGUUG,GUUUUG,UGUGUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CPEB4 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| DDX19B | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | GAAACU,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CELF1 | 5 | 1097 | 0.014285714 | 0.0055320435 | 1.368689 | CUGUCU,GUGUGU,GUGUUG,UGUGUG,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CELF4 | 3 | 776 | 0.009523810 | 0.0039147521 | 1.282618 | GUGUGU,GUGUUG,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUGUGU,GUUUUG,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | AAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUCUU,CUCUUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| HNRNPC | 8 | 2006 | 0.021428571 | 0.0101118501 | 1.083489 | CUUUUU,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | GGAGAU,GGAGGG,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.0069629182 | 1.036809 | CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.