circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000182831:+:16:9102983:9103259
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000182831:+:16:9102983:9103259 | ENSG00000182831 | ENST00000327827 | + | 16 | 9102984 | 9103259 | 276 | AAAGCGUGGAUACCCAUCAACGAAGUUUUGAUAUUGGAAUUCAGAUUGGCUAUCAGCGACGCAAUAAGGAUGUGUUGGCUUGGGUUAAAAAACGCAGAAGAACUAUUCGUCGAGAAGAUUUGAUCAGCUUCCUGUGUGGAAAAGUUCCUCCACCACGAAACUCUAGAGCUCCCCCAAGACUGACUGUAGUGUCCCCUAACCGAGCUACUUCAACGGAAACUAGCUCAUCUGUAGAGACUGAUUUGCAACCCUUCCGGGAAGCCAUAGCUCUGCAUGAAAGCGUGGAUACCCAUCAACGAAGUUUUGAUAUUGGAAUUCAGAUUGGC | circ |
| ENSG00000182831:+:16:9102983:9103259 | ENSG00000182831 | ENST00000327827 | + | 16 | 9102984 | 9103259 | 22 | AGCUCUGCAUGAAAGCGUGGAU | bsj |
| ENSG00000182831:+:16:9102983:9103259 | ENSG00000182831 | ENST00000327827 | + | 16 | 9102784 | 9102993 | 210 | ACUGAAUUUAGACCAUUAAGAUUAAAUGUUACAUAUAACAUUUUCAUUAUGACUGAAAUCAUUUCAGUUUAUAUUACUGAUGGGCUCAGAGGCCACAUCAUGGUAUUCACUUUGGUUAAAUUGUAAAAUGGUUUUCUUUAGAAGGGAAUACGGGCUCUAAUGGUCAUGAUACAUGUUUUCUUUUUCUUUUUUUCUAACAGAAAGCGUGGA | ie_up |
| ENSG00000182831:+:16:9102983:9103259 | ENSG00000182831 | ENST00000327827 | + | 16 | 9103250 | 9103459 | 210 | GCUCUGCAUGGUAAAGCCGUUUAAACAUAUUUCUUUGGAAAAAUGGUUAAGAGGGUGCCUGUGGACCCAUUUUUCACGGUUAGGUUUAGGUCCAGAUAUACUGUUUUUUGUCUUACAGUAAACAGCUUGCUCUAGAAAUGUUUAAUAGAAUGUAAGCUAAGAACUUAAUAUGGGCGAUUUCUGUCAGCUAACCUAUCCUAGAAAAUGUUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
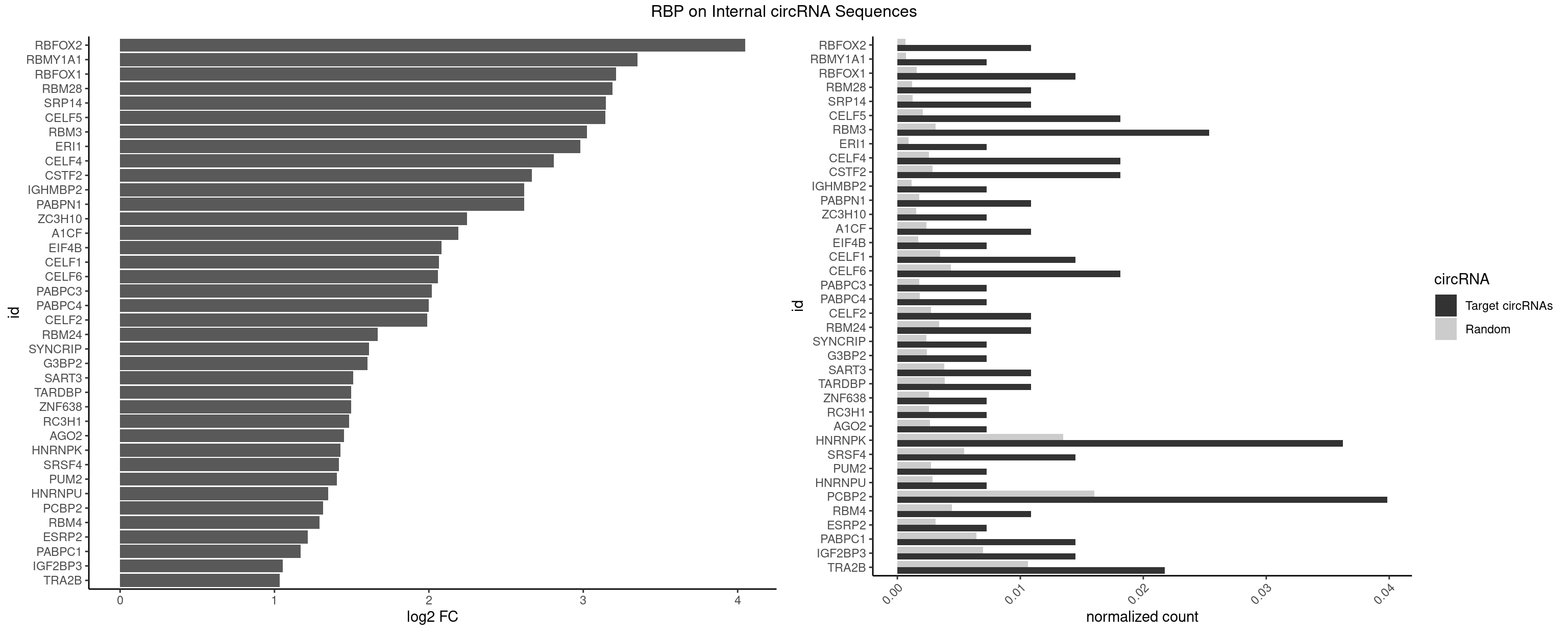
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 2 | 452 | 0.010869565 | 0.0006564894 | 4.049379 | UGACUG,UGCAUG | UGACUG,UGCAUG |
| RBMY1A1 | 1 | 489 | 0.007246377 | 0.0007101099 | 3.351146 | CAAGAC | ACAAGA,CAAGAC |
| RBFOX1 | 3 | 1077 | 0.014492754 | 0.0015622419 | 3.213642 | GCAUGA,UGACUG,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBM28 | 2 | 822 | 0.010869565 | 0.0011926949 | 3.187997 | UGUAGA,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SRP14 | 2 | 847 | 0.010869565 | 0.0012289250 | 3.144826 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| CELF5 | 4 | 1415 | 0.018115942 | 0.0020520728 | 3.142106 | GUGUGG,GUGUUG,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM3 | 6 | 2152 | 0.025362319 | 0.0031201361 | 3.023006 | AAAACG,AAACUA,AAGACU,GAAACU,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ERI1 | 1 | 632 | 0.007246377 | 0.0009173461 | 2.981722 | UUCAGA | UUCAGA,UUUCAG |
| CELF4 | 4 | 1782 | 0.018115942 | 0.0025839306 | 2.809621 | GUGUGG,GUGUUG,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CSTF2 | 4 | 1967 | 0.018115942 | 0.0028520334 | 2.667197 | GUGUUG,GUUUUG,UGUGUG,UGUGUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| IGHMBP2 | 1 | 813 | 0.007246377 | 0.0011796520 | 2.618898 | AAAAAA | AAAAAA |
| PABPN1 | 2 | 1222 | 0.010869565 | 0.0017723764 | 2.616537 | AGAAGA | AAAAGA,AGAAGA |
| ZC3H10 | 1 | 1053 | 0.007246377 | 0.0015274610 | 2.246124 | CAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| A1CF | 2 | 1642 | 0.010869565 | 0.0023810421 | 2.190629 | GAUCAG,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| EIF4B | 1 | 1179 | 0.007246377 | 0.0017100607 | 2.083212 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| CELF1 | 3 | 2391 | 0.014492754 | 0.0034664959 | 2.063782 | GUGUUG,UGUGUG,UGUGUU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CELF6 | 4 | 2997 | 0.018115942 | 0.0043447134 | 2.059927 | GUGUGG,GUGUUG,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| PABPC3 | 1 | 1234 | 0.007246377 | 0.0017897669 | 2.017488 | AAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC4 | 1 | 1251 | 0.007246377 | 0.0018144033 | 1.997765 | AAAAAA | AAAAAA,AAAAAG |
| CELF2 | 2 | 1886 | 0.010869565 | 0.0027346479 | 1.990867 | AUGUGU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM24 | 2 | 2357 | 0.010869565 | 0.0034172229 | 1.669398 | GUGUGG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| SYNCRIP | 1 | 1634 | 0.007246377 | 0.0023694485 | 1.612709 | AAAAAA | AAAAAA,UUUUUU |
| G3BP2 | 1 | 1644 | 0.007246377 | 0.0023839405 | 1.603912 | AGGAUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SART3 | 2 | 2634 | 0.010869565 | 0.0038186524 | 1.509159 | AAAAAA,AAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| TARDBP | 2 | 2654 | 0.010869565 | 0.0038476365 | 1.498250 | GUUUUG,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZNF638 | 1 | 1773 | 0.007246377 | 0.0025708878 | 1.494993 | UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RC3H1 | 1 | 1786 | 0.007246377 | 0.0025897275 | 1.484460 | CCCUUC | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| AGO2 | 1 | 1830 | 0.007246377 | 0.0026534924 | 1.449367 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPK | 9 | 9304 | 0.036231884 | 0.0134848428 | 1.425921 | AAAAAA,ACCCAU,ACCCUU,CAACCC,CCCCAA,CCCCCA,CCCCUA,UCCCCC,UCCCCU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| SRSF4 | 3 | 3740 | 0.014492754 | 0.0054214720 | 1.418575 | AGAAGA,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| PUM2 | 1 | 1890 | 0.007246377 | 0.0027404447 | 1.402850 | UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPU | 1 | 1965 | 0.007246377 | 0.0028491350 | 1.346736 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| PCBP2 | 10 | 11045 | 0.039855072 | 0.0160079069 | 1.315979 | AACCCU,AAUUCA,CCCCCA,CCCUAA,CCCUUC,CCUCCA,CCUUCC,CUAACC,CUCCCC,CUUCCU | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| RBM4 | 2 | 3065 | 0.010869565 | 0.0044432593 | 1.290604 | CCUUCC,CUUCCU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ESRP2 | 1 | 2150 | 0.007246377 | 0.0031172377 | 1.216992 | GGGAAG | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| PABPC1 | 3 | 4443 | 0.014492754 | 0.0064402624 | 1.170140 | AAAAAA,AAAAAC,CUAACC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| IGF2BP3 | 3 | 4815 | 0.014492754 | 0.0069793662 | 1.054164 | AAAAAC,AAACUC,AAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| TRA2B | 5 | 7329 | 0.021739130 | 0.0106226650 | 1.033148 | AAGAAC,AAUAAG,AGAAGA,GAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
RBP on BSJ (Exon Only)
Plot
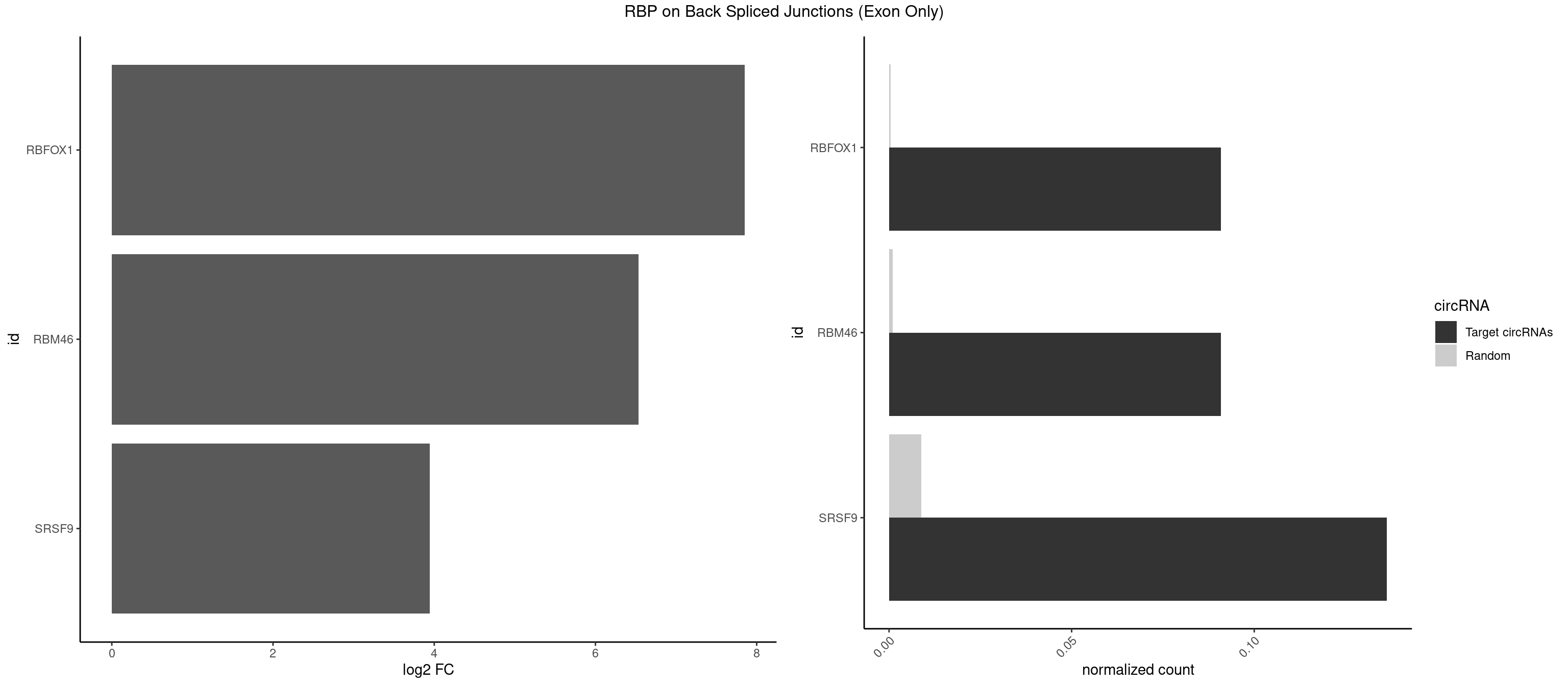
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX1 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GCAUGA | AGCAUG,GCAUGU,UGACUG,UGCAUG |
| RBM46 | 1 | 14 | 0.09090909 | 0.0009824469 | 6.531901 | AUGAAA | AAUCAA,AAUGAU,AUCAAA,AUCAAU,AUGAAG,AUGAUG,AUGAUU,GAUCAU,GAUGAA,GAUGAU |
| SRSF9 | 2 | 134 | 0.13636364 | 0.0088420225 | 3.946939 | AUGAAA,UGAAAG | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
RBP on BSJ (Exon and Intron)
Plot
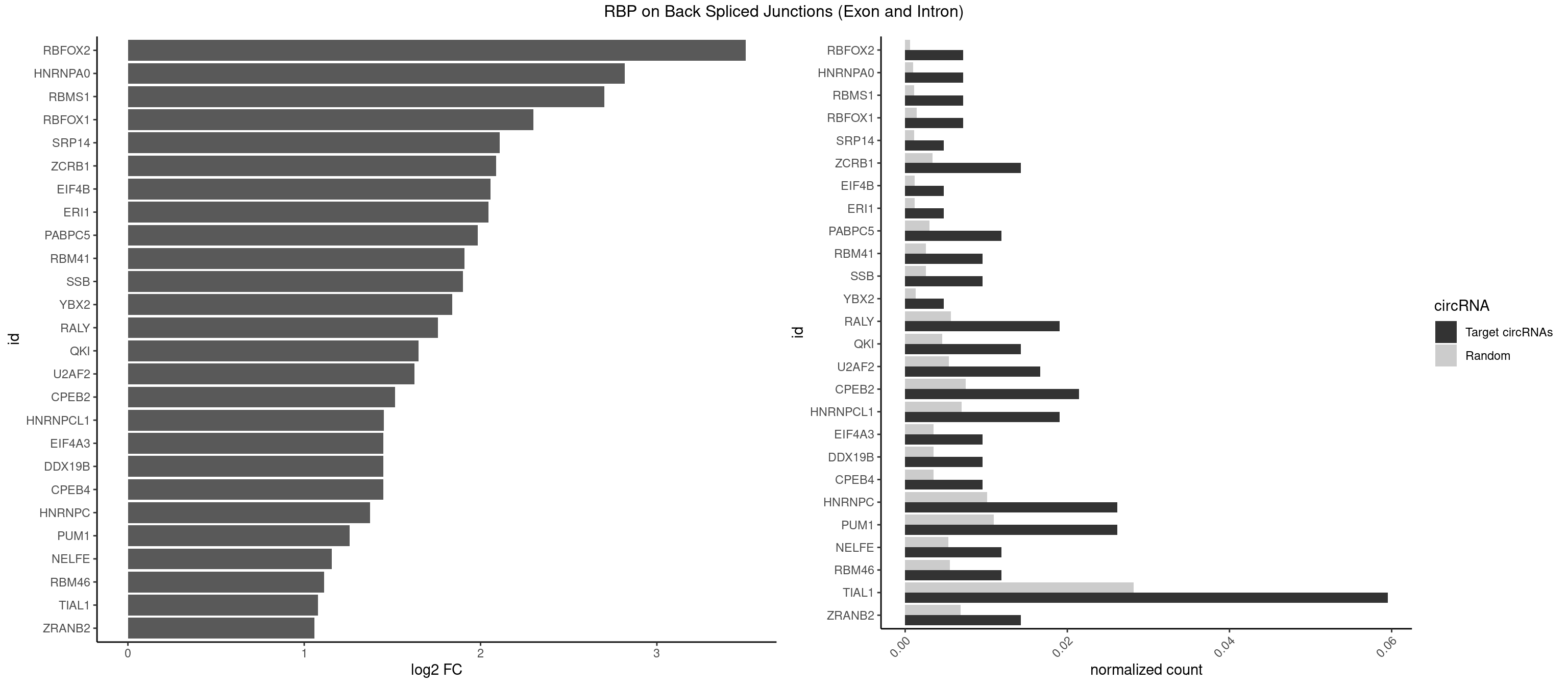
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 2 | 124 | 0.007142857 | 0.0006297864 | 3.503567 | UGACUG,UGCAUG | UGACUG,UGCAUG |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMS1 | 2 | 217 | 0.007142857 | 0.0010983474 | 2.701167 | AUAUAC,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | UGACUG,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ZCRB1 | 5 | 666 | 0.014285714 | 0.0033605401 | 2.087808 | ACUUAA,GAUUAA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| PABPC5 | 4 | 596 | 0.011904762 | 0.0030078597 | 1.984730 | AGAAAA,AGAAAG,AGAAAU,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | AUACAU,UACAUG,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RALY | 7 | 1117 | 0.019047619 | 0.0056328094 | 1.757684 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | AAUCAU,CUAACA,CUAACC,UAACCU,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| U2AF2 | 6 | 1071 | 0.016666667 | 0.0054010480 | 1.625654 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| CPEB2 | 8 | 1487 | 0.021428571 | 0.0074969770 | 1.515155 | AUUUUU,CAUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPCL1 | 7 | 1381 | 0.019047619 | 0.0069629182 | 1.451847 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB4 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| DDX19B | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| HNRNPC | 10 | 2006 | 0.026190476 | 0.0101118501 | 1.372995 | AUUUUU,CUUUUU,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PUM1 | 10 | 2172 | 0.026190476 | 0.0109482064 | 1.258348 | AAUGUU,AAUUGU,AUUGUA,GUCCAG,UAAUAU,UACAUA,UGUAAA,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | GCUAAC,GGUUAG,UGGUUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUCAU,AUCAUG,AUCAUU,AUGAUA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| TIAL1 | 24 | 5597 | 0.059523810 | 0.0282043531 | 1.077549 | AUUUUC,AUUUUU,CAGUUU,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UCAGUU,UUAAAU,UUCAGU,UUUAAA,UUUCAG,UUUUCA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZRANB2 | 5 | 1360 | 0.014285714 | 0.0068571141 | 1.058900 | AGGGAA,AGGUUU,GGUAAA,GUAAAG,UGGUAA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.