circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000109919:-:11:47625673:47627127
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000109919:-:11:47625673:47627127 | ENSG00000109919 | ENST00000302503 | - | 11 | 47625674 | 47627127 | 116 | GUUUCUACCAUGAAUGAAAUGAAGAGUUAUUCUCAAGCUGUCACAGGAUUUUUUGCGAGUAUGUUGACCUAUCCCUUUGUGCUUGUCUCCAAUCUUAUGGCUGUCAACAACUGUGGGUUUCUACCAUGAAUGAAAUGAAGAGUUAUUCUCAAGCUGUCACAGGAUU | circ |
| ENSG00000109919:-:11:47625673:47627127 | ENSG00000109919 | ENST00000302503 | - | 11 | 47625674 | 47627127 | 22 | AACAACUGUGGGUUUCUACCAU | bsj |
| ENSG00000109919:-:11:47625673:47627127 | ENSG00000109919 | ENST00000302503 | - | 11 | 47627118 | 47627327 | 210 | AGACACCAUUUCUUAAAAAAUAUUUUGAAAUUAGCUGGGCGUGGUGAUGUGUGCUUCAGGAGGUCACUGCACUUCAGCCUGGGUGACAGAGUGAGGCACUGUCUGAAAGAAAAAAAGAAAAGAAAAGAAAAGGAAAAAAGAAUAUAUUGAUGUGUUGUAGUGUUGUAGGUAAUUAAUUUAAGGCUCUUUUCCUAUUUUAGGUUUCUACCA | ie_up |
| ENSG00000109919:-:11:47625673:47627127 | ENSG00000109919 | ENST00000302503 | - | 11 | 47625474 | 47625683 | 210 | ACAACUGUGGGUAAGCACUUUGUAUUUCUUAUUCUAGUAGGUGGUUGACUGUAUGCUGACAGGCGGAGCAGCCUUGUAGAAAUUUAGGAAAGAUGAAUGAUCAGUAUCAAUCUCUGAUUGGUAUUUUUUUAAAAAGGGGGGGAGGAAAAGUAUAAUUGAUACCAUUAGAAGAUUCCUAAUUUUUUUCUUUGUAUGCCCUAUGCAGCAGCA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
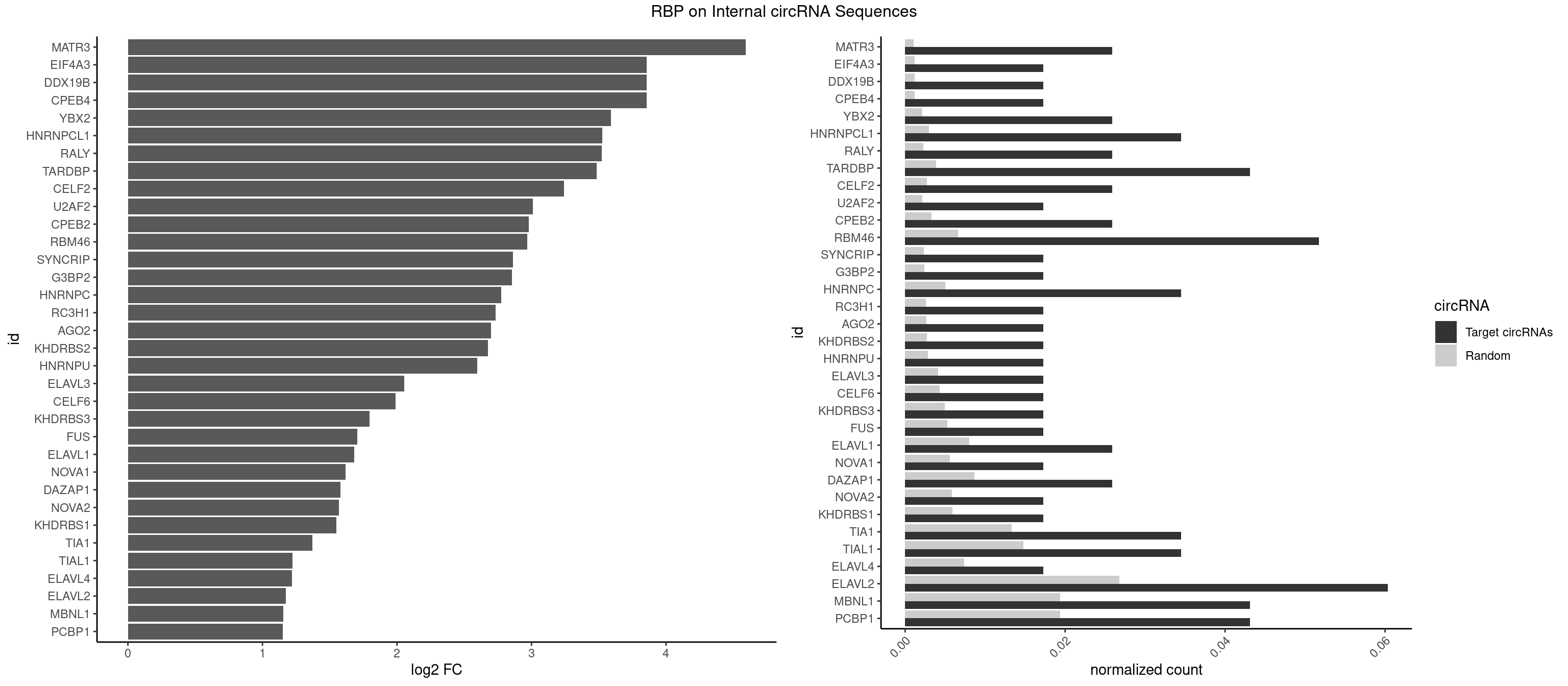
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MATR3 | 2 | 739 | 0.02586207 | 0.001072411 | 4.591908 | AAUCUU,AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| CPEB4 | 1 | 821 | 0.01724138 | 0.001191246 | 3.855332 | UUUUUU | UUUUUU |
| DDX19B | 1 | 821 | 0.01724138 | 0.001191246 | 3.855332 | UUUUUU | UUUUUU |
| EIF4A3 | 1 | 821 | 0.01724138 | 0.001191246 | 3.855332 | UUUUUU | UUUUUU |
| YBX2 | 2 | 1480 | 0.02586207 | 0.002146271 | 3.590934 | AACAAC,ACAACU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| HNRNPCL1 | 3 | 2062 | 0.03448276 | 0.002989708 | 3.527799 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| RALY | 2 | 1553 | 0.02586207 | 0.002252063 | 3.521519 | UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| TARDBP | 4 | 2654 | 0.04310345 | 0.003847636 | 3.485759 | GAAUGA,UGAAUG,UUGUGC,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| CELF2 | 2 | 1886 | 0.02586207 | 0.002734648 | 3.241411 | GUAUGU,UAUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| U2AF2 | 1 | 1477 | 0.01724138 | 0.002141923 | 3.008896 | UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| CPEB2 | 2 | 2261 | 0.02586207 | 0.003278099 | 2.979906 | AUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RBM46 | 5 | 4554 | 0.05172414 | 0.006601124 | 2.970054 | AAUGAA,AUGAAA,AUGAAG,AUGAAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SYNCRIP | 1 | 1634 | 0.01724138 | 0.002369448 | 2.863252 | UUUUUU | AAAAAA,UUUUUU |
| G3BP2 | 1 | 1644 | 0.01724138 | 0.002383941 | 2.854455 | AGGAUU | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPC | 3 | 3471 | 0.03448276 | 0.005031636 | 2.776776 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RC3H1 | 1 | 1786 | 0.01724138 | 0.002589727 | 2.735003 | UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| AGO2 | 1 | 1830 | 0.01724138 | 0.002653492 | 2.699911 | GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| KHDRBS2 | 1 | 1858 | 0.01724138 | 0.002694070 | 2.678016 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPU | 1 | 1965 | 0.01724138 | 0.002849135 | 2.597279 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| ELAVL3 | 1 | 2867 | 0.01724138 | 0.004156317 | 2.052498 | UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| CELF6 | 1 | 2997 | 0.01724138 | 0.004344713 | 1.988542 | UGUGGG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| KHDRBS3 | 1 | 3429 | 0.01724138 | 0.004970770 | 1.794334 | UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| FUS | 1 | 3648 | 0.01724138 | 0.005288145 | 1.705041 | UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ELAVL1 | 2 | 5554 | 0.02586207 | 0.008050328 | 1.683718 | UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| NOVA1 | 1 | 3872 | 0.01724138 | 0.005612767 | 1.619091 | UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| DAZAP1 | 2 | 5964 | 0.02586207 | 0.008644502 | 1.580983 | AGUAUG,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| NOVA2 | 1 | 4013 | 0.01724138 | 0.005817105 | 1.567502 | UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS1 | 1 | 4064 | 0.01724138 | 0.005891014 | 1.549287 | UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| TIA1 | 3 | 9206 | 0.03448276 | 0.013342821 | 1.369811 | AUUUUU,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIAL1 | 3 | 10182 | 0.03448276 | 0.014757244 | 1.224452 | AUUUUU,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL4 | 1 | 5105 | 0.01724138 | 0.007399635 | 1.220349 | UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| ELAVL2 | 6 | 18468 | 0.06034483 | 0.026765348 | 1.172864 | AUUUUU,GAUUUU,UAUGUU,UCUUAU,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| MBNL1 | 4 | 13372 | 0.04310345 | 0.019380204 | 1.153219 | GCUUGU,GUGCUU,UGUCUC,UUGUGC | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| PCBP1 | 4 | 13384 | 0.04310345 | 0.019397595 | 1.151926 | CCAAUC,CCUAUC,CUAUCC,UUUUUU | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
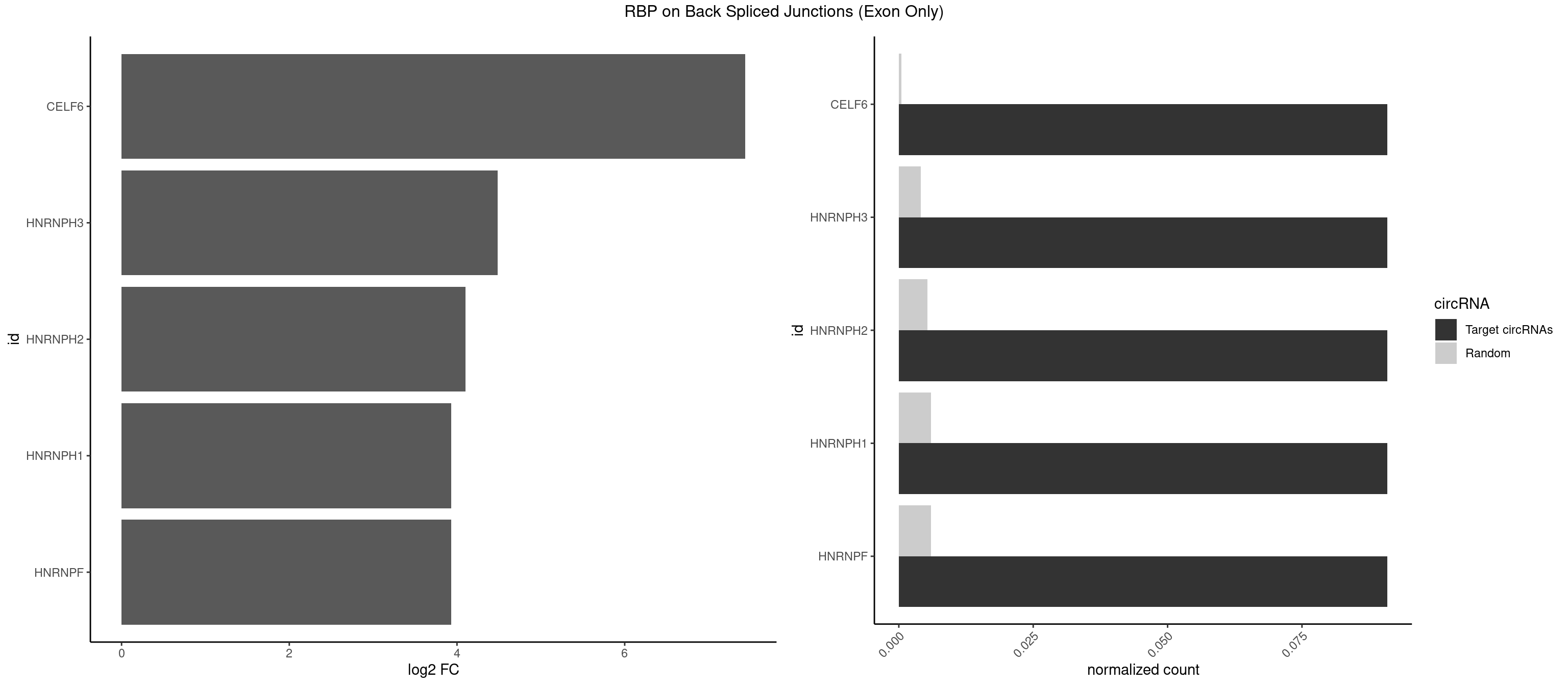
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CELF6 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | UGUGGG | GUGAGG,GUGUUG,UGUGGG,UGUGUG |
| HNRNPH3 | 1 | 61 | 0.09090909 | 0.0040607807 | 4.484596 | UGUGGG | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| HNRNPH2 | 1 | 80 | 0.09090909 | 0.0053052135 | 4.098942 | UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPF | 1 | 90 | 0.09090909 | 0.0059601782 | 3.930997 | UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 1 | 90 | 0.09090909 | 0.0059601782 | 3.930997 | UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
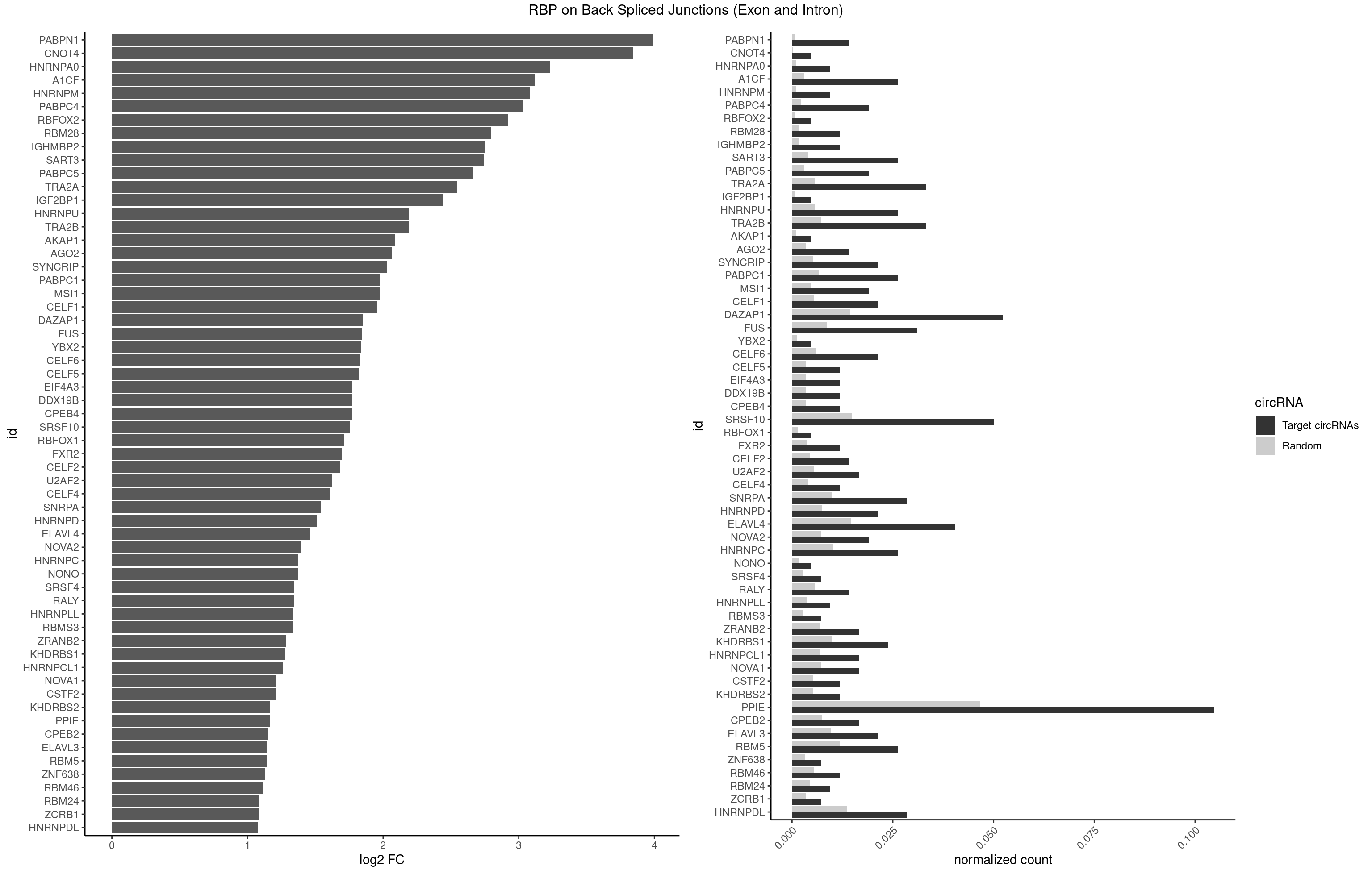
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PABPN1 | 5 | 178 | 0.014285714 | 0.0009018541 | 3.985535 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| HNRNPA0 | 3 | 200 | 0.009523810 | 0.0010126965 | 3.233337 | AAUUUA,AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| A1CF | 10 | 598 | 0.026190476 | 0.0030179363 | 3.117408 | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPM | 3 | 222 | 0.009523810 | 0.0011235389 | 3.083489 | AAGGAA,GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| PABPC4 | 7 | 462 | 0.019047619 | 0.0023327287 | 3.029520 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| RBM28 | 4 | 340 | 0.011904762 | 0.0017180572 | 2.792689 | AGUAGG,UGUAGA,UGUAGG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| IGHMBP2 | 4 | 350 | 0.011904762 | 0.0017684401 | 2.750989 | AAAAAA | AAAAAA |
| SART3 | 10 | 777 | 0.026190476 | 0.0039197904 | 2.740194 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPC5 | 7 | 596 | 0.019047619 | 0.0030078597 | 2.662801 | AGAAAA,AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TRA2A | 13 | 1133 | 0.033333333 | 0.0057134220 | 2.544539 | AAAGAA,AAGAAA,AGAAGA,GAAAGA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPU | 10 | 1136 | 0.026190476 | 0.0057285369 | 2.192804 | AAAAAA,GGGGGG,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| TRA2B | 13 | 1447 | 0.033333333 | 0.0072954454 | 2.191898 | AAAGAA,AAGAAU,AAGGAA,AGAAGA,AGGAAA,GAAAGA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| AGO2 | 5 | 677 | 0.014285714 | 0.0034159613 | 2.064210 | AAAAAA,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SYNCRIP | 8 | 1041 | 0.021428571 | 0.0052498992 | 2.029174 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| PABPC1 | 10 | 1321 | 0.026190476 | 0.0066606207 | 1.975314 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| MSI1 | 7 | 961 | 0.019047619 | 0.0048468360 | 1.974496 | AGGAGG,AGGUGG,AGUAGG,UAGGAA,UAGGUA,UAGGUG,UAGUAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| CELF1 | 8 | 1097 | 0.021428571 | 0.0055320435 | 1.953651 | CUGUCU,GUGUUG,UGUCUG,UGUGUG,UGUGUU,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| DAZAP1 | 21 | 2878 | 0.052380952 | 0.0145052398 | 1.852468 | AAAAAA,AAUUUA,AGGAAA,AGGUAA,AGUAGG,AGUAUA,GGGGGG,UAGGAA,UAGGUA,UAGGUU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| FUS | 12 | 1711 | 0.030952381 | 0.0086255542 | 1.843361 | AAAAAA,GGGGGG,GGGUGA,UGGUGA,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAACU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF6 | 8 | 1196 | 0.021428571 | 0.0060308343 | 1.829106 | GUGAGG,GUGAUG,GUGGUG,GUGUUG,UGUGGG,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUUG,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CPEB4 | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| DDX19B | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| EIF4A3 | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| SRSF10 | 20 | 2937 | 0.050000000 | 0.0148024990 | 1.756087 | AAAAGA,AAAGAA,AAAGGA,AAAGGG,AAGAAA,AAGGAA,AAGGGG,GAAAGA,GAGGAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| FXR2 | 4 | 730 | 0.011904762 | 0.0036829907 | 1.692589 | GACAGA,GACAGG,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| CELF2 | 5 | 881 | 0.014285714 | 0.0044437727 | 1.684716 | AUGUGU,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| U2AF2 | 6 | 1071 | 0.016666667 | 0.0054010480 | 1.625654 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GUGUUG,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SNRPA | 11 | 1947 | 0.028571429 | 0.0098145909 | 1.541573 | AGUAGG,AUGCUG,AUUCCU,GAGCAG,GAUACC,GAUUCC,GUAGUG,GUAUGC,UAUGCU,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| HNRNPD | 8 | 1488 | 0.021428571 | 0.0075020153 | 1.514186 | AAAAAA,AAUUUA,AGUAGG,UUAGGA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ELAVL4 | 16 | 2916 | 0.040476190 | 0.0146966949 | 1.461582 | AAAAAA,UAUUUU,UUGUAU,UUUGUA,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| NOVA2 | 7 | 1434 | 0.019047619 | 0.0072299476 | 1.397554 | GAGGCA,GGGGGG,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| HNRNPC | 10 | 2006 | 0.026190476 | 0.0101118501 | 1.372995 | AUUUUU,GGGGGG,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RALY | 5 | 1117 | 0.014285714 | 0.0056328094 | 1.342647 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AGAAGA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPLL | 3 | 748 | 0.009523810 | 0.0037736800 | 1.335567 | ACACCA,ACUGCA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | AAUAUA,AUAUAU | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ZRANB2 | 6 | 1360 | 0.016666667 | 0.0068571141 | 1.281292 | AGGUAA,AGGUUU,GGGUAA,GGGUGA,GGUGGU,GUGGUG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| KHDRBS1 | 9 | 1945 | 0.023809524 | 0.0098045143 | 1.280021 | AUUUAA,UAAAAA,UUAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| HNRNPCL1 | 6 | 1381 | 0.016666667 | 0.0069629182 | 1.259202 | AUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| NOVA1 | 6 | 1428 | 0.016666667 | 0.0071997179 | 1.210953 | GGGGGG,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUUG,UGUGUG,UGUGUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| PPIE | 43 | 9262 | 0.104761905 | 0.0466696896 | 1.166556 | AAAAAA,AAAAAU,AAAAUA,AAAUAU,AAAUUA,AAAUUU,AAUAUA,AAUAUU,AAUUAA,AAUUUA,AAUUUU,AUAAUU,AUAUAU,AUAUUU,AUUAAU,AUUUAA,AUUUUA,AUUUUU,UAAAAA,UAAUUA,UAAUUU,UAUAAU,UAUAUU,UAUUUU,UUAAAA,UUAAUU,UUUAAA,UUUUAA,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| CPEB2 | 6 | 1487 | 0.016666667 | 0.0074969770 | 1.152585 | AUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ELAVL3 | 8 | 1928 | 0.021428571 | 0.0097188634 | 1.140676 | AUUUUA,UAUUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| RBM5 | 10 | 2359 | 0.026190476 | 0.0118903668 | 1.139249 | AAAAAA,AAGGAA,AAGGGG,AGGUAA,GGGGGG,GGUGGU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUGAU,AUCAAU,AUGAAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | AGAGUG,GAGUGA,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA,AUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPDL | 11 | 2689 | 0.028571429 | 0.0135530028 | 1.075961 | AAUUUA,ACACCA,ACUGCA,AGUAGG,AUACCA,AUUAGC,CAUUAG,CUAGUA,UUUAGG | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.