circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000163938:+:3:52687245:52688192
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000163938:+:3:52687245:52688192 | ENSG00000163938 | ENST00000496254 | + | 3 | 52687246 | 52688192 | 336 | GUUCGAGAACAUCAUCGAAAAUUAAGAAAGGAGGCUAAAAAGCGGGGUCACAAGAAGCCUAGGAAAGACCCAGGAGUUCCAAACAGUGCUCCCUUUAAGGAGGCUCUUCUUAGGGAAGCUGAGCUAAGGAAACAGAGGCUUGAAGAACUAAAACAGCAGCAGAAACUUGACAGGCAGAAGGAACUAGAAAAGAAAAGAAAACUUGAAACUAAUCCUGAUAUUAAGCCAUCAAAUGUGGAACCUAUGGAAAAGGAGUUUGGGCUUUGCAAAACUGAGAACAAAGCCAAGUCGGGCAAACAGAAUUCAAAGAAGCUGUACUGCCAAGAACUUAAAAAGGUUCGAGAACAUCAUCGAAAAUUAAGAAAGGAGGCUAAAAAGCGGGGUCA | circ |
| ENSG00000163938:+:3:52687245:52688192 | ENSG00000163938 | ENST00000496254 | + | 3 | 52687246 | 52688192 | 22 | AACUUAAAAAGGUUCGAGAACA | bsj |
| ENSG00000163938:+:3:52687245:52688192 | ENSG00000163938 | ENST00000496254 | + | 3 | 52687046 | 52687255 | 210 | GCAUAGAGAGCCUUCUAGUUACAAUUUCUUGCUGUUUUAUACUUACAUAUGCAUUACUUUGUAAGAUUCCAAUUAAAGCUCCAUUUUCCUAGGACAUUUUAUAGGCAUAACUAAAUUGCAGCCAGAUUGGUUUCUCACUUGAAUUCUGCUUAAGUAUAAAGAUAUUUUUGUAAGCAGACAAAAUCUCUUUAUUUUAAUAGGUUCGAGAAC | ie_up |
| ENSG00000163938:+:3:52687245:52688192 | ENSG00000163938 | ENST00000496254 | + | 3 | 52688183 | 52688392 | 210 | ACUUAAAAAGGUAUCUUAGCCUAGGUCAGUGUCUGACAGUAGUAAUGAGGUUUAAAAGACUCAAGUCAUUUUUUUUUUAACCUUUUAAGAUGAAGGGUGCAUGUGCAGGUUUGUUAUAUGGGUAAACUUGCGUCAUAGGGGUUUGCUGUACAGAUUAUUUCAUCACCCAGGUAUUAAGCCUAGUACGACAUUAGUUAUCUUUCCUGAUCC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
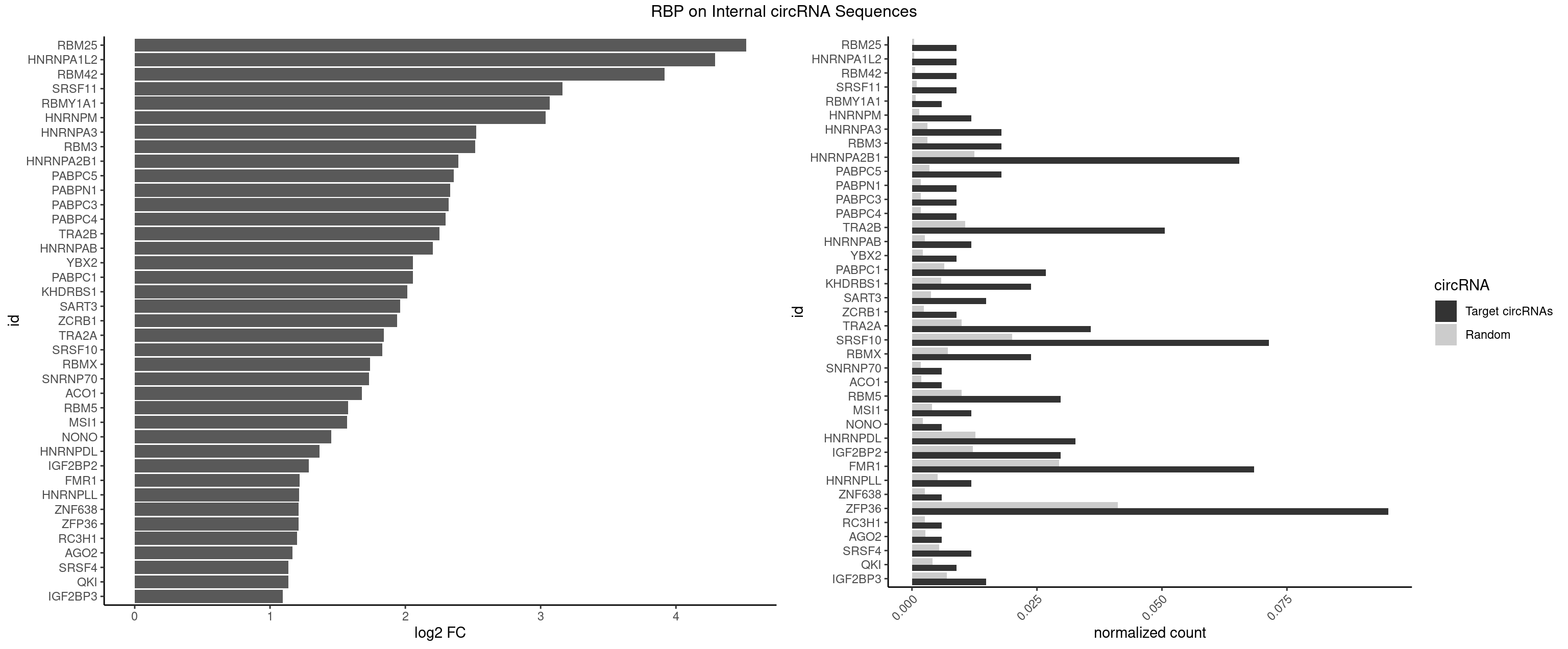
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 2 | 268 | 0.008928571 | 0.0003898359 | 4.517491 | CGGGCA,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| HNRNPA1L2 | 2 | 314 | 0.008928571 | 0.0004564992 | 4.289745 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBM42 | 2 | 407 | 0.008928571 | 0.0005912752 | 3.916528 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| SRSF11 | 2 | 688 | 0.008928571 | 0.0009985015 | 3.160593 | AAGAAG | AAGAAG |
| RBMY1A1 | 1 | 489 | 0.005952381 | 0.0007101099 | 3.067353 | ACAAGA | ACAAGA,CAAGAC |
| HNRNPM | 3 | 999 | 0.011904762 | 0.0014492040 | 3.038206 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| HNRNPA3 | 5 | 2140 | 0.017857143 | 0.0031027457 | 2.524884 | AAGGAG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM3 | 5 | 2152 | 0.017857143 | 0.0031201361 | 2.516820 | AAAACU,AAACUA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPA2B1 | 21 | 8607 | 0.065476190 | 0.0124747476 | 2.391960 | AAGAAG,AAGGAA,AAGGAG,ACUAGA,AGAAGC,AGGAAC,CAAGAA,CCAAGA,GAAGCC,GAAGGA,GCCAAG,GGAACC,UAGGGA,UUAGGG | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| PABPC5 | 5 | 2400 | 0.017857143 | 0.0034795387 | 2.359533 | AGAAAA,AGAAAG,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PABPN1 | 2 | 1222 | 0.008928571 | 0.0017723764 | 2.332744 | AAAAGA | AAAAGA,AGAAGA |
| PABPC3 | 2 | 1234 | 0.008928571 | 0.0017897669 | 2.318658 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC4 | 2 | 1251 | 0.008928571 | 0.0018144033 | 2.298934 | AAAAAG | AAAAAA,AAAAAG |
| TRA2B | 16 | 7329 | 0.050595238 | 0.0106226650 | 2.251856 | AAAGAA,AAGAAC,AAGAAG,AAGGAA,AGAAGG,AGGAAA,GAAAGA,GAAGAA,GAAGGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| HNRNPAB | 3 | 1782 | 0.011904762 | 0.0025839306 | 2.203900 | AAAGAC,ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| YBX2 | 2 | 1480 | 0.008928571 | 0.0021462711 | 2.056597 | AACAUC,ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PABPC1 | 8 | 4443 | 0.026785714 | 0.0064402624 | 2.056272 | ACUAAU,AGAAAA,CAAACA,CUAAUC,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| KHDRBS1 | 7 | 4064 | 0.023809524 | 0.0058910141 | 2.014951 | CUAAAA,GAAAAC,UAAAAA,UAAAAC,UUAAAA | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| SART3 | 4 | 2634 | 0.014880952 | 0.0038186524 | 1.962331 | AGAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ZCRB1 | 2 | 1605 | 0.008928571 | 0.0023274215 | 1.939697 | AAUUAA,ACUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| TRA2A | 11 | 6871 | 0.035714286 | 0.0099589296 | 1.842439 | AAAGAA,AAGAAA,AAGAAG,AGAAAG,GAAAGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SRSF10 | 23 | 13860 | 0.071428571 | 0.0200874160 | 1.830209 | AAAAGA,AAAGAA,AAAGAC,AAAGGA,AAGAAA,AAGAAG,AAGGAA,AAGGAG,ACAAAG,CAAAGA,GAAAGA,GAGAAC | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| RBMX | 7 | 4925 | 0.023809524 | 0.0071387787 | 1.737790 | AAGAAG,AAGGAA,AGAAGG,AUCAAA,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SNRNP70 | 1 | 1237 | 0.005952381 | 0.0017941145 | 1.730195 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ACO1 | 1 | 1283 | 0.005952381 | 0.0018607779 | 1.677561 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM5 | 9 | 6879 | 0.029761905 | 0.0099705232 | 1.577726 | AAGGAA,AAGGAG,AGGGAA,CUCUUC,GAAGGA,UCUUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| MSI1 | 3 | 2770 | 0.011904762 | 0.0040157442 | 1.567800 | AGGAGG,UAGGAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| NONO | 1 | 1498 | 0.005952381 | 0.0021723567 | 1.454206 | AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| HNRNPDL | 10 | 8759 | 0.032738095 | 0.0126950266 | 1.366707 | AACAGC,AACUAA,ACAGCA,ACUAGA,CCUUUA,CUAGGA,CUUUAA,GAACUA | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| IGF2BP2 | 9 | 8408 | 0.029761905 | 0.0121863560 | 1.288200 | AAAACA,AAUUCA,CAAAAC,CAAACA,CCAAAC,GAAAAC,GAAUUC,GCAAAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| FMR1 | 22 | 20291 | 0.068452381 | 0.0294072466 | 1.218929 | AAGCGG,AAGGAA,AAGGAG,AGCAGC,AGGAGG,AGGAGU,CUAAGG,CUAUGG,GAAGGA,GACAGG,GCUAAG,GCUGAG,GGCUAA,UAAGGA,UAUGGA,UGACAG | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| HNRNPLL | 3 | 3534 | 0.011904762 | 0.0051229360 | 1.216496 | CAAACA,GCAAAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| ZNF638 | 1 | 1773 | 0.005952381 | 0.0025708878 | 1.211200 | GGUUCG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ZFP36 | 31 | 28400 | 0.095238095 | 0.0411588414 | 1.210336 | AAAAAG,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAGAA,AAAGGA,AACAAA,AAGAAA,AAGGAA,ACAAAG,AGAAAG,AGGAAA,CAAACA,CAAAGA,CCCUUU,GGAAAG,UAAGGA,UCUUCU | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
| RC3H1 | 1 | 1786 | 0.005952381 | 0.0025897275 | 1.200667 | UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| AGO2 | 1 | 1830 | 0.005952381 | 0.0026534924 | 1.165574 | AGUGCU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SRSF4 | 3 | 3740 | 0.011904762 | 0.0054214720 | 1.134782 | AAGAAG,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| QKI | 2 | 2805 | 0.008928571 | 0.0040664663 | 1.134654 | ACUAAU,CUAAUC | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| IGF2BP3 | 4 | 4815 | 0.014880952 | 0.0069793662 | 1.092299 | AAAACA,AAUUCA,CAAACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
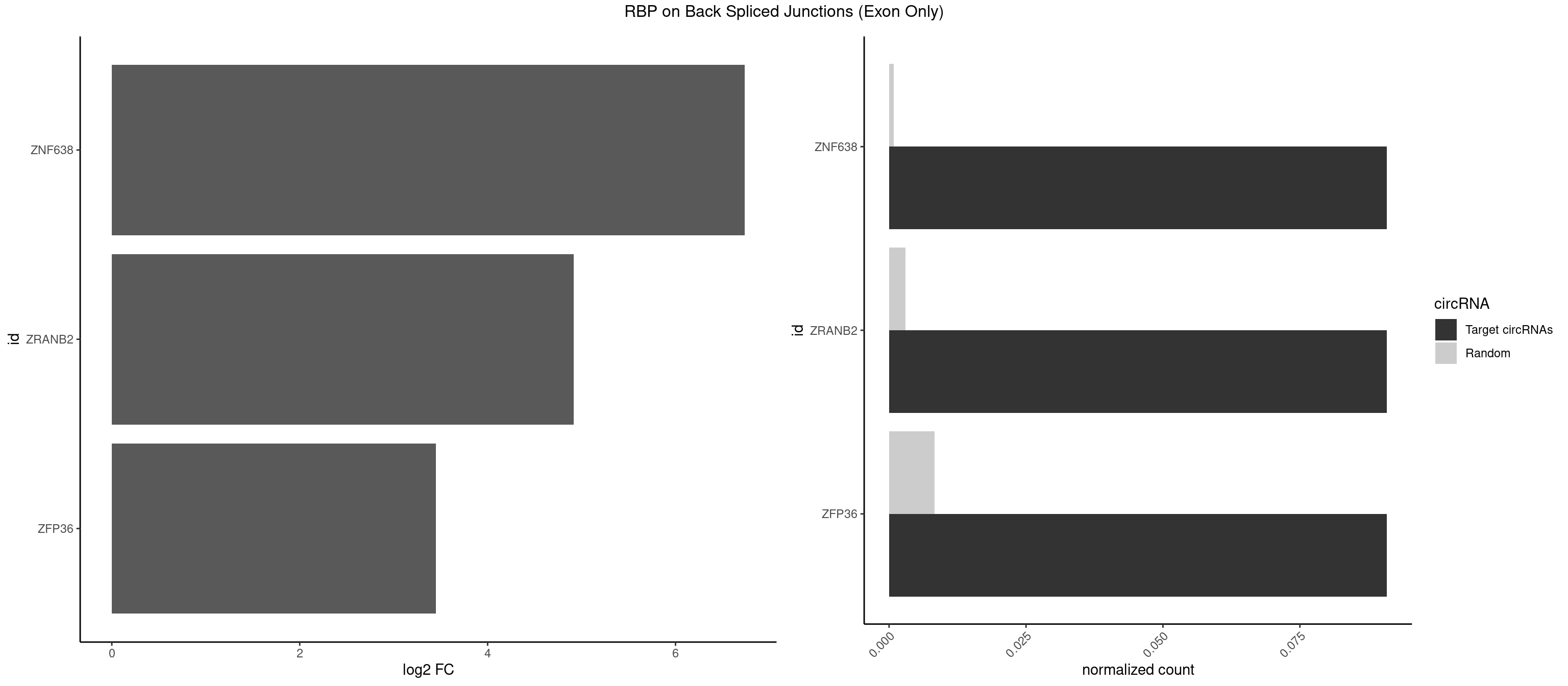
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZNF638 | 1 | 12 | 0.09090909 | 0.000851454 | 6.738352 | GGUUCG | CGUUCG,CGUUCU,CGUUGG,GGUUCU,GGUUGG,GUUCUU,GUUGGU,UGUUCU,UGUUGG,UGUUGU |
| ZRANB2 | 1 | 45 | 0.09090909 | 0.003012837 | 4.915230 | AAAGGU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.008318051 | 3.450107 | AAAAGG | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
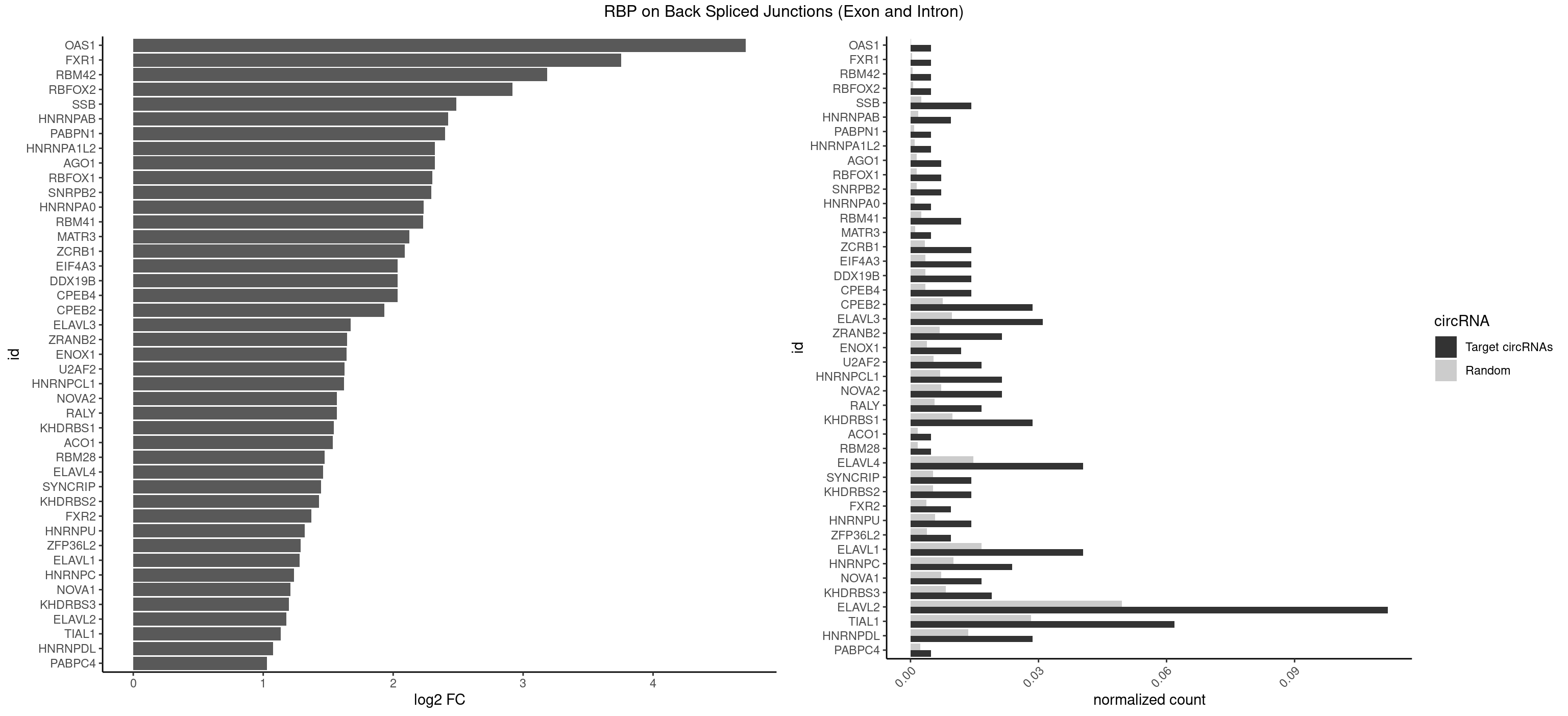
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 35 | 0.004761905 | 0.0001813785 | 4.714464 | GCAUAA | GCAUAA |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | ACGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| SSB | 5 | 505 | 0.014285714 | 0.0025493753 | 2.486358 | CUGUUU,GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPAB | 3 | 351 | 0.009523810 | 0.0017734784 | 2.424957 | AAAGAC,AGACAA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | AUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | GUAGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | AUUGCA,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | AUACUU,UACUUU,UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ZCRB1 | 5 | 666 | 0.014285714 | 0.0033605401 | 2.087808 | AAUUAA,ACUUAA,GCUUAA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CPEB4 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| DDX19B | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| CPEB2 | 11 | 1487 | 0.028571429 | 0.0074969770 | 1.930192 | AUUUUU,CAUUUU,CCUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ELAVL3 | 12 | 1928 | 0.030952381 | 0.0097188634 | 1.671191 | AUUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ZRANB2 | 8 | 1360 | 0.021428571 | 0.0068571141 | 1.643862 | AAAGGU,AGGUAU,AGGUUU,GAGGUU,GGGUAA,GGUAAA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ENOX1 | 4 | 756 | 0.011904762 | 0.0038139863 | 1.642167 | AGGACA,CAGACA,GUACAG,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| U2AF2 | 6 | 1071 | 0.016666667 | 0.0054010480 | 1.625654 | UUUUCC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| HNRNPCL1 | 8 | 1381 | 0.021428571 | 0.0069629182 | 1.621772 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| NOVA2 | 8 | 1434 | 0.021428571 | 0.0072299476 | 1.567479 | AGGCAU,AGUCAU,AUCACC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| RALY | 6 | 1117 | 0.016666667 | 0.0056328094 | 1.565039 | UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 | 11 | 1945 | 0.028571429 | 0.0098045143 | 1.543055 | CAAAAU,CUAAAU,UAAAAA,UAAAAG,UUAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ELAVL4 | 16 | 2916 | 0.040476190 | 0.0146966949 | 1.461582 | GUAUCU,UAUUUU,UUAUUU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| SYNCRIP | 5 | 1041 | 0.014285714 | 0.0052498992 | 1.444212 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 5 | 1051 | 0.014285714 | 0.0053002821 | 1.430432 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| FXR2 | 3 | 730 | 0.009523810 | 0.0036829907 | 1.370661 | AGACAA,GACAAA,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPU | 5 | 1136 | 0.014285714 | 0.0057285369 | 1.318335 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.0039046755 | 1.286336 | UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| ELAVL1 | 16 | 3309 | 0.040476190 | 0.0166767432 | 1.279236 | UAUUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGGUU,UUUAUU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| HNRNPC | 9 | 2006 | 0.023809524 | 0.0101118501 | 1.235492 | AUUUUU,UUUUUA,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| NOVA1 | 6 | 1428 | 0.016666667 | 0.0071997179 | 1.210953 | AUCACC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| KHDRBS3 | 7 | 1646 | 0.019047619 | 0.0082980653 | 1.198764 | AUAAAG,AUUAAA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ELAVL2 | 46 | 9826 | 0.111904762 | 0.0495112858 | 1.176442 | AUACUU,AUAUUU,AUUAUU,AUUUUA,AUUUUC,AUUUUU,CAUUUU,CUUUUA,GUUUUA,UACUUU,UAGUUA,UAUACU,UAUUUU,UCAUUU,UUAAGU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUUU,UUUAAG,UUUAAU,UUUAUA,UUUAUU,UUUCAU,UUUCUU,UUUUAA,UUUUAU,UUUUUA,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIAL1 | 25 | 5597 | 0.061904762 | 0.0282043531 | 1.134133 | AUUUUA,AUUUUC,AUUUUU,CUUUUA,UAAAUU,UAUUUU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPDL | 11 | 2689 | 0.028571429 | 0.0135530028 | 1.075961 | AACCUU,AACUAA,ACAUUA,AGAUAU,CAUUAG,CUAGGA,CUAGUA,UAAGUA,UAGGCA,UCUGAC,UUAGCC | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.