circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000101752:+:18:21765771:21779685
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000101752:+:18:21765771:21779685 | ENSG00000101752 | ENST00000261537 | + | 18 | 21765772 | 21779685 | 679 | GCAUCAAGCAUGAUGGAACCAUGUGUGAUACCUGCCGCCAGCAACCAAUCAUUGGCAUUCGAUGGAAGUGUGCAGAGUGUACAAAUUAUGAUUUGUGCACAGUGUGUUAUCAUGGAGAUAAACAUCAUUUAAGACAUCGCUUUUACCGAAUUACUACACCGGGAAGUGAGAGGGUUCUGUUAGAGUCUCGUAGGAAAUCUAAGAAGAUUACAGCCAGAGGAAUCUUUGCAGGUGCCAGAGUGGUGCGAGGAGUGGACUGGCAGUGGGAAGAUCAAGAUGGAGGAAAUGGACGUAGGGGAAAGGUAACAGAAAUCCAGGACUGGAGUGCAUCAAGCCCACAUAGCGCAGCAUAUGUCCUCUGGGAUAAUGGUGCUAAGAACCUUUACAGAGUUGGCUUUGAGGGCAUGUCUGAUCUGAAAUGUGUCCAGGAUGCCAAGGGAGGUUCUUUCUACAGAGAUCACUGCCCUGUGCUAGGUGAGCAGAAUGGCAACAGGAAUCCUGGUGGAUUGCAGAUUGGUGACCUGGUAAAUAUAGAUCUCGACCUCGAAAUUGUACAGUCUUUGCAGCAUGGUCAUGGAGGAUGGACUGAUGGAAUGUUUGAGACUUUAACUACAACUGGAACUGUUUGUGGCAUUGAUGAAGAUCAUGACAUUGUAGUACAGUAUCCAAGUGGCAAUAGGCAUCAAGCAUGAUGGAACCAUGUGUGAUACCUGCCGCCAGCAACCAAUC | circ |
| ENSG00000101752:+:18:21765771:21779685 | ENSG00000101752 | ENST00000261537 | + | 18 | 21765772 | 21779685 | 22 | AGUGGCAAUAGGCAUCAAGCAU | bsj |
| ENSG00000101752:+:18:21765771:21779685 | ENSG00000101752 | ENST00000261537 | + | 18 | 21765572 | 21765781 | 210 | GGAAAAUAUAUAUAAUUCGCAUUUAUUAACGUAGUUGAGAUCCAUUAUCUUGAAGCUAUAUGUGUUAAAUUUGAAGUGUUAUUUCCUAAUUUAUUAGAAUAUCUUGUAAAAUGUUCUGUUCUUAUUUUUUUCCCCCAAGGAAUAAUAUUCAAAUAAAUAAAAUUUGUGAUUAAUCUGAGCAUGUGUCCUUGUUUUAAUAGGCAUCAAGCA | ie_up |
| ENSG00000101752:+:18:21765771:21779685 | ENSG00000101752 | ENST00000261537 | + | 18 | 21779676 | 21779885 | 210 | GUGGCAAUAGGUGGGUGAUAUCUCUCAAUUUUUGACAAUGACAAAUAAAACUAAUAUAGAGAAAAGUCUUAGAGAAAGCAAAUAAAGUGAUACAACCAAAUACUCAUUAAAGCUAGCUUUUAGAUGGUAAGUAAAAAUCCAGAAUAUAUAUUGGUAAGGUGGAGAGAAACAAAUGAUAAGAAUUACGUGUGUGUGUGUGUGUGUGUGUGU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
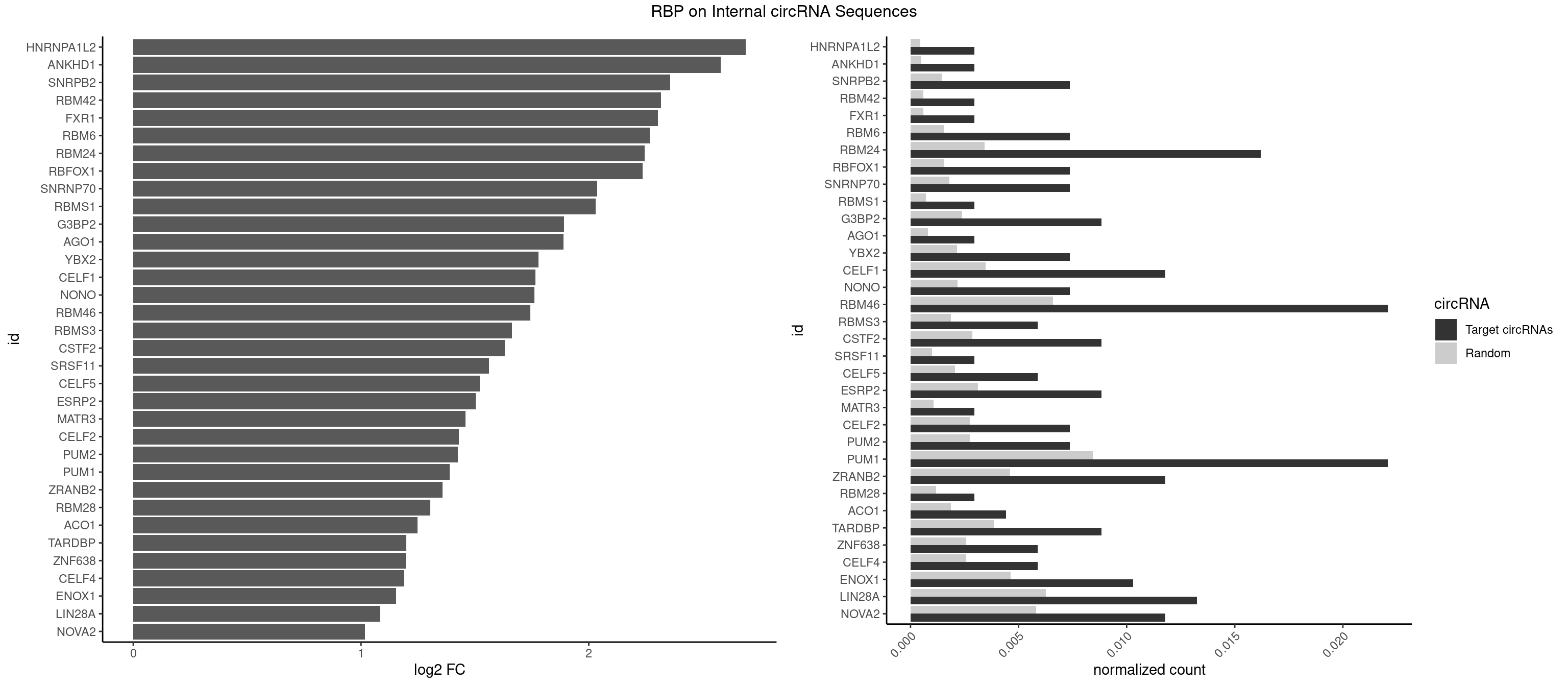
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 1 | 314 | 0.002945508 | 0.0004564992 | 2.689832 | GUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| ANKHD1 | 1 | 339 | 0.002945508 | 0.0004927293 | 2.579649 | GACGUA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SNRPB2 | 4 | 991 | 0.007363770 | 0.0014376103 | 2.356772 | AUUGCA,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM42 | 1 | 407 | 0.002945508 | 0.0005912752 | 2.316615 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR1 | 1 | 411 | 0.002945508 | 0.0005970720 | 2.302540 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM6 | 4 | 1054 | 0.007363770 | 0.0015289102 | 2.267941 | AAUCCA,AUCCAA,AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBM24 | 10 | 2357 | 0.016200295 | 0.0034172229 | 2.245124 | AGAGUG,AGUGUG,GAGUGG,GAGUGU,GUGUGA,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| RBFOX1 | 4 | 1077 | 0.007363770 | 0.0015622419 | 2.236827 | AGCAUG,GCAUGA,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRNP70 | 4 | 1237 | 0.007363770 | 0.0017941145 | 2.037173 | AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBMS1 | 1 | 497 | 0.002945508 | 0.0007217036 | 2.029038 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| G3BP2 | 5 | 1644 | 0.008836524 | 0.0023839405 | 1.890131 | AGGAUG,GGAUAA,GGAUGG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| AGO1 | 1 | 548 | 0.002945508 | 0.0007956130 | 1.888378 | GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| YBX2 | 4 | 1480 | 0.007363770 | 0.0021462711 | 1.778612 | AACAUC,ACAACU,ACAUCA,ACAUCG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF1 | 7 | 2391 | 0.011782032 | 0.0034664959 | 1.765038 | GUGUGU,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| NONO | 4 | 1498 | 0.007363770 | 0.0021723567 | 1.761184 | AGAGGA,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM46 | 14 | 4554 | 0.022091311 | 0.0066011240 | 1.742695 | AAUCAU,AUCAAG,AUCAUG,AUCAUU,AUGAAG,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBMS3 | 3 | 1283 | 0.005891016 | 0.0018607779 | 1.662611 | AAUAUA,AUAUAG,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| CSTF2 | 5 | 1967 | 0.008836524 | 0.0028520334 | 1.631488 | GUGUGU,UGUGUG,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| SRSF11 | 1 | 688 | 0.002945508 | 0.0009985015 | 1.560680 | AAGAAG | AAGAAG |
| CELF5 | 3 | 1415 | 0.005891016 | 0.0020520728 | 1.521435 | GUGUGU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ESRP2 | 5 | 2150 | 0.008836524 | 0.0031172377 | 1.503211 | GGGAAA,GGGAAG,GGGGAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| MATR3 | 1 | 739 | 0.002945508 | 0.0010724109 | 1.457659 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| CELF2 | 4 | 1886 | 0.007363770 | 0.0027346479 | 1.429090 | AUGUGU,GUGUGU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PUM2 | 4 | 1890 | 0.007363770 | 0.0027404447 | 1.426035 | GUAAAU,UAAAUA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PUM1 | 14 | 5823 | 0.022091311 | 0.0084401638 | 1.388136 | AAUGUU,AAUUGU,AGAUAA,AUUGUA,CAGAAU,GUAAAU,GUCCAG,UAAAUA,UGUACA,UGUCCA,UUGUAC,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ZRANB2 | 7 | 3173 | 0.011782032 | 0.0045997733 | 1.356954 | AAAGGU,AGGUAA,GAGGUU,GGUAAA,GUGGUG,UGGUAA,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| RBM28 | 1 | 822 | 0.002945508 | 0.0011926949 | 1.304292 | UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ACO1 | 2 | 1283 | 0.004418262 | 0.0018607779 | 1.247573 | CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| TARDBP | 5 | 2654 | 0.008836524 | 0.0038476365 | 1.199507 | GAAUGG,GAAUGU,GUGUGU,UGUGUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZNF638 | 3 | 1773 | 0.005891016 | 0.0025708878 | 1.196250 | GGUUCU,GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF4 | 3 | 1782 | 0.005891016 | 0.0025839306 | 1.188949 | GUGUGU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ENOX1 | 6 | 3195 | 0.010309278 | 0.0046316558 | 1.154343 | AAGACA,AGUACA,GUACAG,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| LIN28A | 8 | 4315 | 0.013254786 | 0.0062547643 | 1.083486 | AGGAGU,GGAGAU,GGAGGA,UGGAGA,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| NOVA2 | 7 | 4013 | 0.011782032 | 0.0058171047 | 1.018215 | AGACAU,AGAUCA,AGGCAU,GAUCAC,UAGAUC | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
RBP on BSJ (Exon Only)
Plot
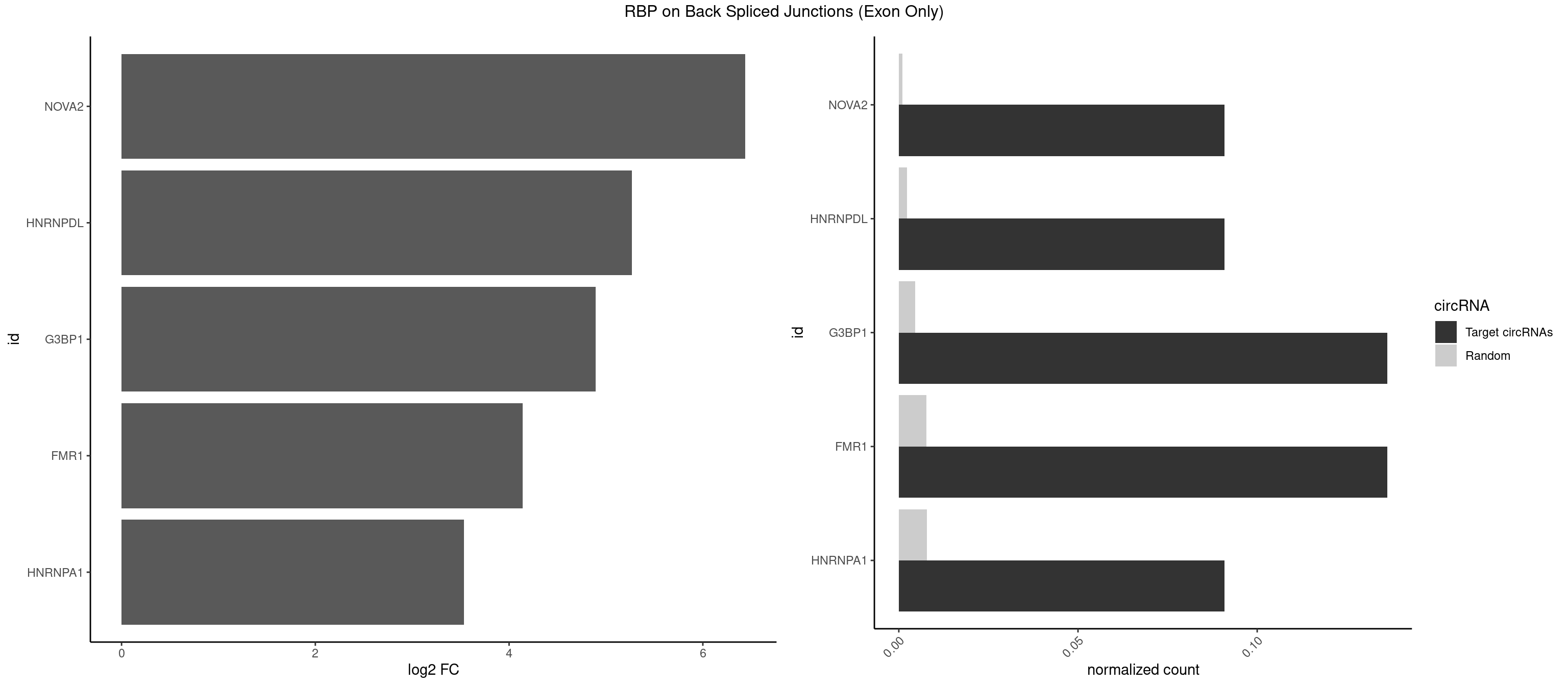
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NOVA2 | 1 | 15 | 0.09090909 | 0.001047943 | 6.438792 | AGGCAU | ACCACC,AGACAU,AGAUCA,AGGCAU,AGUCAU,CUAGAU,GAGACA,GAGGCA,GAGUCA,UAGAUC |
| HNRNPDL | 1 | 35 | 0.09090909 | 0.002357873 | 5.268867 | UAGGCA | AACAGC,AAUACC,AAUUUA,ACACCA,ACAGCA,ACCAGA,AGAUAU,AGUAGG,AUCUGA,AUUAGC,AUUAGG,CACGCA,CCAGAC,CUAAGC,CUAAGU,CUAGAU,CUAGGA,CUAGGC,CUUUAG,GAACUA,GAUUAG,GCACUA,GCUAGU,UGCGCA,UUAGCC,UUAGGC,UUUAGG |
| G3BP1 | 2 | 69 | 0.13636364 | 0.004584752 | 4.894471 | AUAGGC,UAGGCA | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| FMR1 | 2 | 117 | 0.13636364 | 0.007728583 | 4.141111 | AUAGGC,UAGGCA | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.007859576 | 3.531901 | UAGGCA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
RBP on BSJ (Exon and Intron)
Plot
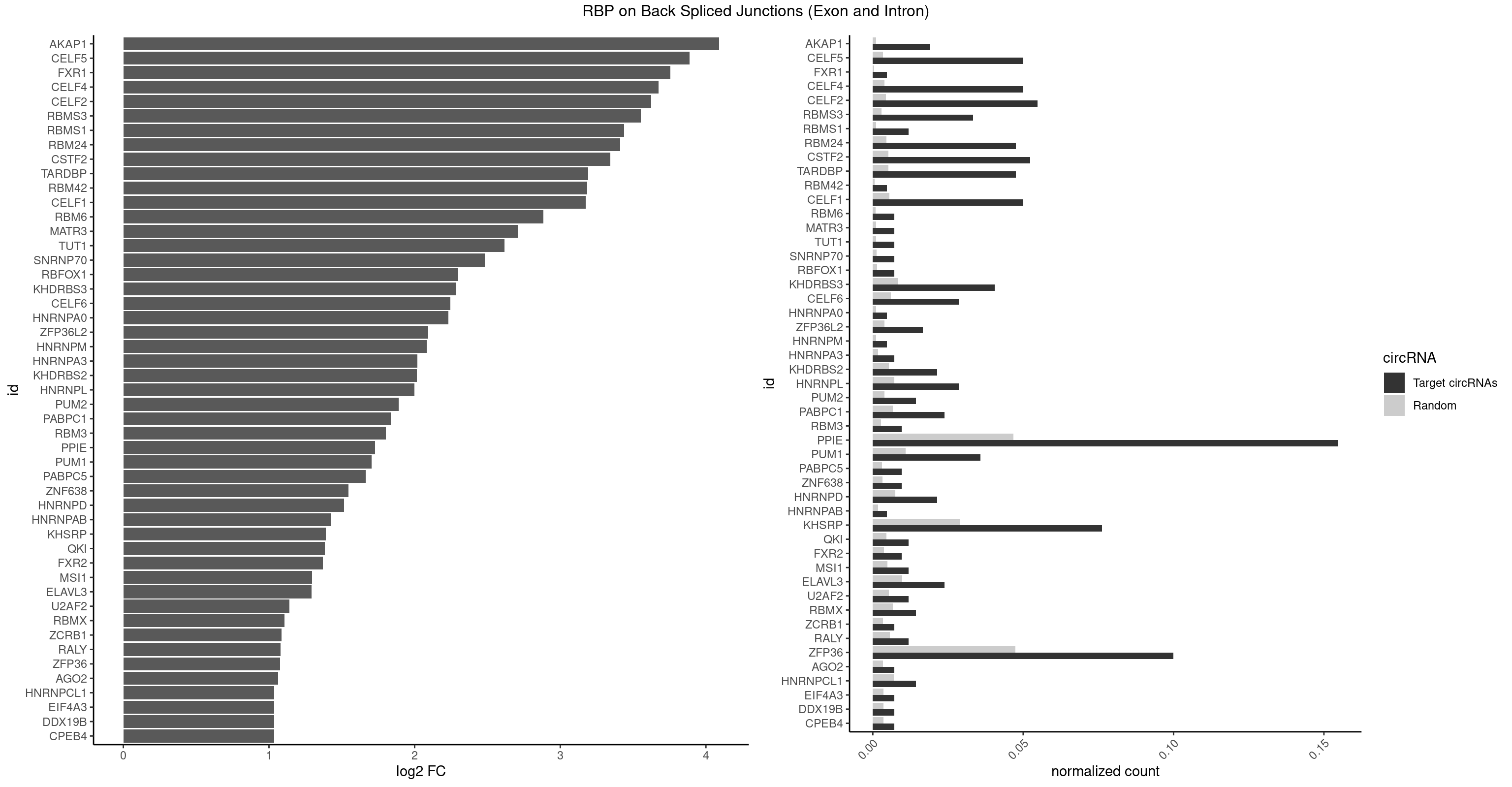
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| AKAP1 | 7 | 221 | 0.019047619 | 0.0011185006 | 4.089973 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| CELF5 | 20 | 669 | 0.050000000 | 0.0033756550 | 3.888689 | GUGUGU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| CELF4 | 20 | 776 | 0.050000000 | 0.0039147521 | 3.674935 | GUGUGU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF2 | 22 | 881 | 0.054761905 | 0.0044437727 | 3.623316 | AUGUGU,GUGUGU,UAUGUG,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBMS3 | 13 | 562 | 0.033333333 | 0.0028365578 | 3.554752 | AAUAUA,AUAUAG,AUAUAU,CUAUAU,UAUAGA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBMS1 | 4 | 217 | 0.011904762 | 0.0010983474 | 3.438132 | AUAUAG,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBM24 | 19 | 888 | 0.047619048 | 0.0044790407 | 3.410277 | GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CSTF2 | 21 | 1022 | 0.052380952 | 0.0051541717 | 3.345230 | GUGUGU,UGUGUG,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 19 | 1034 | 0.047619048 | 0.0052146312 | 3.190902 | GUGUGU,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| CELF1 | 20 | 1097 | 0.050000000 | 0.0055320435 | 3.176044 | GUGUGU,UGUGUG,UGUGUU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AUCAAG,AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | AGCAUG,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| KHDRBS3 | 16 | 1646 | 0.040476190 | 0.0082980653 | 2.286227 | AAAUAA,AAUAAA,AUAAAA,AUAAAG,AUUAAA,UAAAAC,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| CELF6 | 11 | 1196 | 0.028571429 | 0.0060308343 | 2.244144 | UGUGAU,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| ZFP36L2 | 6 | 774 | 0.016666667 | 0.0039046755 | 2.093691 | AUUUAU,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | CAAGGA,CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| KHDRBS2 | 8 | 1051 | 0.021428571 | 0.0053002821 | 2.015395 | AAUAAA,AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPL | 11 | 1419 | 0.028571429 | 0.0071543732 | 1.997676 | AAACAA,AAAUAA,AAAUAC,AACAAA,AAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| PUM2 | 5 | 764 | 0.014285714 | 0.0038542926 | 1.890035 | UAAAUA,UAUAUA,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC1 | 9 | 1321 | 0.023809524 | 0.0066606207 | 1.837810 | ACAAAU,ACUAAU,AGAAAA,CAAAUA,CUAAUA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AAACUA,AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PPIE | 64 | 9262 | 0.154761905 | 0.0466696896 | 1.729493 | AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUUAAA,AUUAAU,AUUUAU,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAU,UAAUUU,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUU,UUAAAU,UUAAUA,UUAUUA,UUAUUU,UUUAAU,UUUAUU,UUUUAA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| PUM1 | 14 | 2172 | 0.035714286 | 0.0109482064 | 1.705807 | AAUAUU,AAUGUU,AGAAUU,CAGAAU,CCAGAA,CUUGUA,UAAAUA,UAAUAU,UAUAUA,UGUAAA,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAA,AGAAAG,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GUUCUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPD | 8 | 1488 | 0.021428571 | 0.0075020153 | 1.514186 | AAUUUA,AUUUAU,UUAGAG,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| KHSRP | 31 | 5764 | 0.076190476 | 0.0290457477 | 1.391284 | AUGUGU,AUUUAU,GUGUGU,UAUAUU,UAUUUU,UGUGUG,UUAUUA,UUAUUU,UUUAUU | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACUAAU,ACUCAU,AUUAAC,UACUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| FXR2 | 3 | 730 | 0.009523810 | 0.0036829907 | 1.370661 | GACAAA,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGUGG,UAGGUG,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ELAVL3 | 9 | 1928 | 0.023809524 | 0.0097188634 | 1.292679 | AUUUAU,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RBMX | 5 | 1316 | 0.014285714 | 0.0066354293 | 1.106311 | AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGUGUU | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAAUUA,GAUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ZFP36 | 41 | 9408 | 0.100000000 | 0.0474052801 | 1.076880 | AAAAAU,AAAAGU,AAAAUA,AAACAA,AAAGCA,AAAUAA,AACAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,ACAAAU,AGAAAG,AGCAAA,AGUAAA,AUAAAG,AUAAAU,AUUUAU,CAAAUA,CAAGCA,CAAGGA,CCUUGU,UAAAUA,UAAGUA,UCUUGU,UUAUUU,UUUAAU,UUUAUU | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.0069629182 | 1.036809 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.