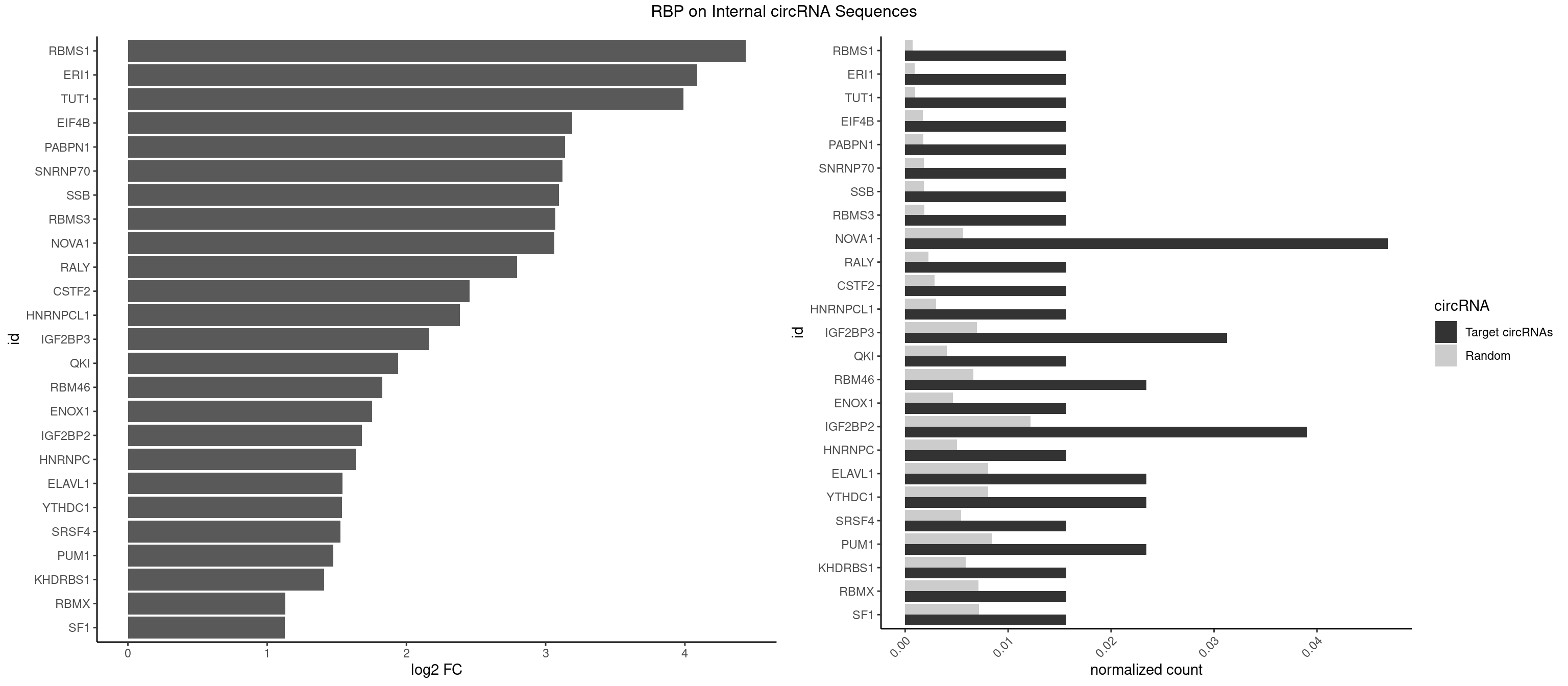
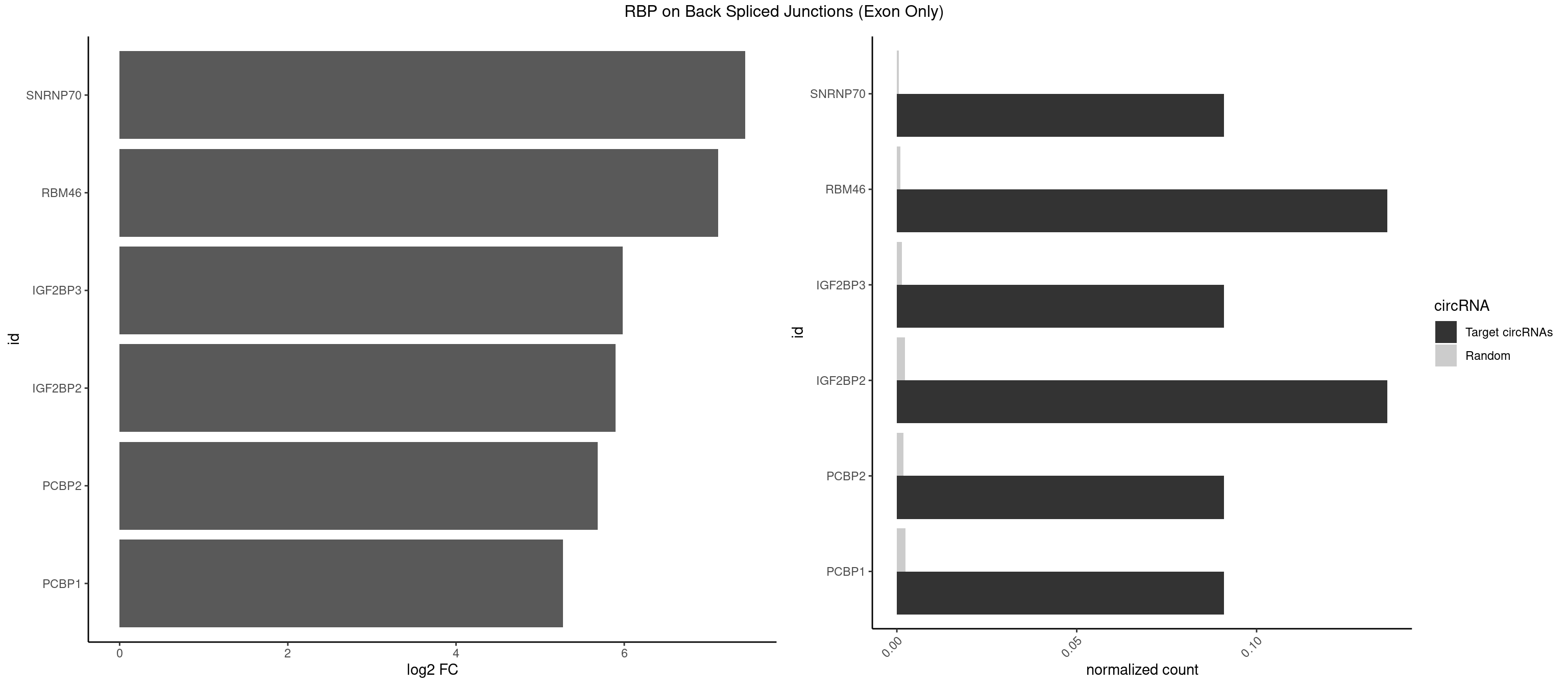
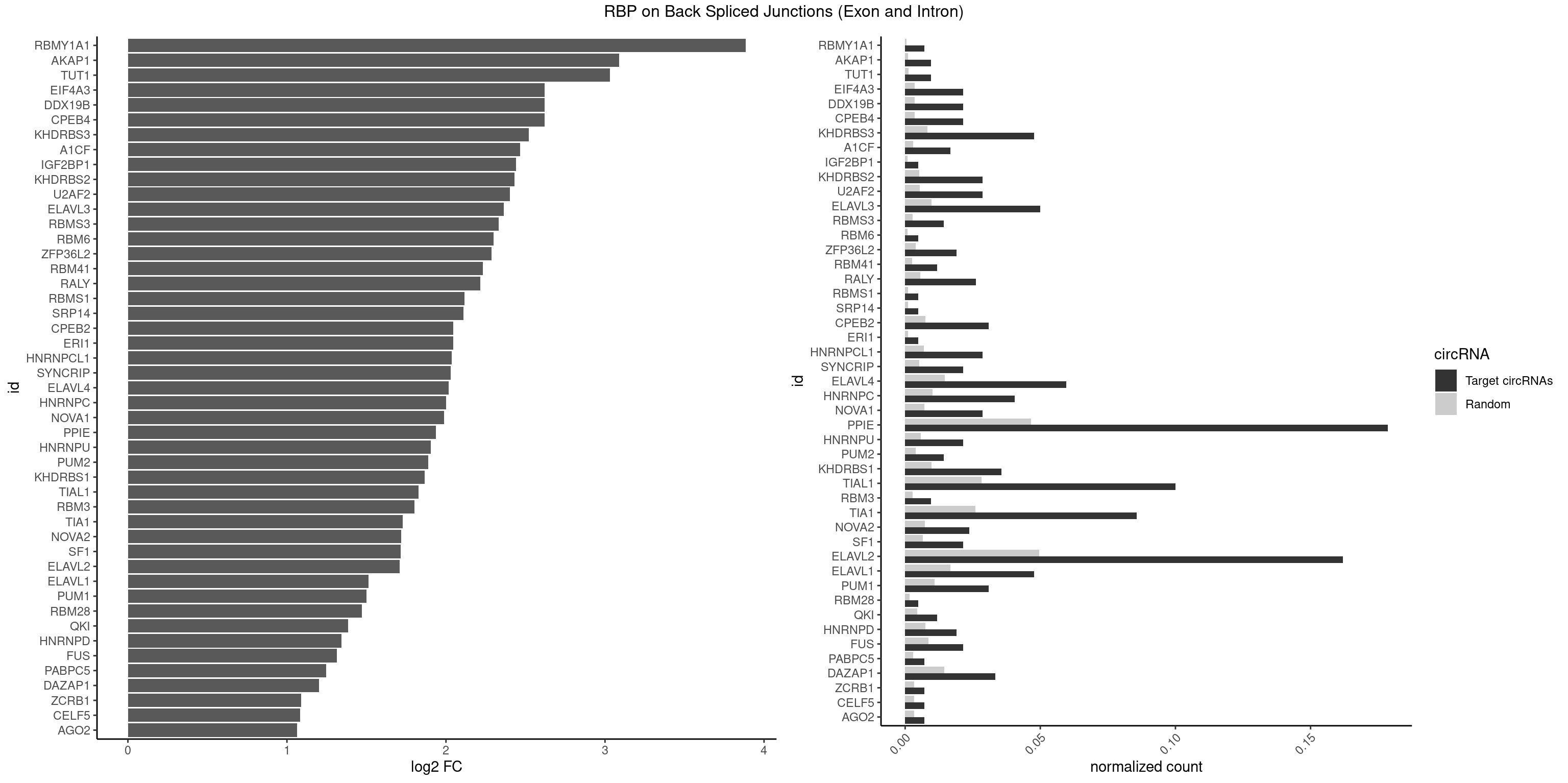
| RBMY1A1 |
2 |
95 |
0.007142857 |
0.0004836759 |
3.884389 |
ACAAGA |
ACAAGA,CAAGAC |
| AKAP1 |
3 |
221 |
0.009523810 |
0.0011185006 |
3.089973 |
AUAUAU,UAUAUA |
AUAUAU,UAUAUA |
| TUT1 |
3 |
230 |
0.009523810 |
0.0011638452 |
3.032640 |
AAAUAC,AAUACU,AGAUAC |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| CPEB4 |
8 |
691 |
0.021428571 |
0.0034864974 |
2.619685 |
UUUUUU |
UUUUUU |
| DDX19B |
8 |
691 |
0.021428571 |
0.0034864974 |
2.619685 |
UUUUUU |
UUUUUU |
| EIF4A3 |
8 |
691 |
0.021428571 |
0.0034864974 |
2.619685 |
UUUUUU |
UUUUUU |
| KHDRBS3 |
19 |
1646 |
0.047619048 |
0.0082980653 |
2.520692 |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAG,UAAAUA,UUAAAC,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| A1CF |
6 |
598 |
0.016666667 |
0.0030179363 |
2.465331 |
AGUAUA,AUAAUU,AUCAGU,UCAGUA,UUAAUU |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| IGF2BP1 |
1 |
173 |
0.004761905 |
0.0008766626 |
2.441445 |
AGCACC |
AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| KHDRBS2 |
11 |
1051 |
0.028571429 |
0.0053002821 |
2.430432 |
AAUAAA,AUAAAA,UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| U2AF2 |
11 |
1071 |
0.028571429 |
0.0054010480 |
2.403262 |
UUUUCC,UUUUUC,UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| ELAVL3 |
20 |
1928 |
0.050000000 |
0.0097188634 |
2.363069 |
AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| RBMS3 |
5 |
562 |
0.014285714 |
0.0028365578 |
2.332360 |
AAUAUA,AUAUAU,CUAUAU,UAUAUA |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBM6 |
1 |
191 |
0.004761905 |
0.0009673519 |
2.299426 |
AAUCCA |
AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ZFP36L2 |
7 |
774 |
0.019047619 |
0.0039046755 |
2.286336 |
UAUUUA,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM41 |
4 |
502 |
0.011904762 |
0.0025342604 |
2.231902 |
AUACUU,UACAUU,UACUUU,UUACUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RALY |
10 |
1117 |
0.026190476 |
0.0056328094 |
2.217116 |
UUUUUC,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| RBMS1 |
1 |
217 |
0.004761905 |
0.0010983474 |
2.116204 |
UAUAUA |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SRP14 |
1 |
218 |
0.004761905 |
0.0011033857 |
2.109602 |
CUGUAG |
CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| CPEB2 |
12 |
1487 |
0.030952381 |
0.0074969770 |
2.045670 |
AUUUUU,CAUUUU,CUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ERI1 |
1 |
228 |
0.004761905 |
0.0011537686 |
2.045185 |
UUCAGA |
UUCAGA,UUUCAG |
| HNRNPCL1 |
11 |
1381 |
0.028571429 |
0.0069629182 |
2.036809 |
AUUUUU,CUUUUU,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SYNCRIP |
8 |
1041 |
0.021428571 |
0.0052498992 |
2.029174 |
UUUUUU |
AAAAAA,UUUUUU |
| ELAVL4 |
24 |
2916 |
0.059523810 |
0.0146966949 |
2.017975 |
UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPC |
16 |
2006 |
0.040476190 |
0.0101118501 |
2.001027 |
AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| NOVA1 |
11 |
1428 |
0.028571429 |
0.0071997179 |
1.988561 |
AGCACC,AUUCAU,UUCAUU,UUUUUU |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| PPIE |
74 |
9262 |
0.178571429 |
0.0466696896 |
1.935943 |
AAAAAU,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAUAAA,AAUAAU,AAUAUA,AAUUAA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAU,AUAUUU,AUUAAU,AUUAUU,AUUUAA,AUUUUA,AUUUUU,UAAAAU,UAAAUA,UAAUUU,UAUAAA,UAUAUA,UAUAUU,UAUUUA,UAUUUU,UUAAAU,UUAAUU,UUAUAA,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPU |
8 |
1136 |
0.021428571 |
0.0057285369 |
1.903297 |
UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| PUM2 |
5 |
764 |
0.014285714 |
0.0038542926 |
1.890035 |
UAAAUA,UAUAUA,UGUAAA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| KHDRBS1 |
14 |
1945 |
0.035714286 |
0.0098045143 |
1.864983 |
AUAAAA,AUUUAA,AUUUAC,CUAAAU,UUAAAU,UUUUAC,UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| TIAL1 |
41 |
5597 |
0.100000000 |
0.0282043531 |
1.826010 |
AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUU,CUUUUA,CUUUUG,CUUUUU,GUUUUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM3 |
3 |
541 |
0.009523810 |
0.0027307537 |
1.802240 |
AAACUA,AAUACU,AUACUA |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| TIA1 |
35 |
5140 |
0.085714286 |
0.0259018541 |
1.726480 |
AUUUUC,AUUUUU,CUUUUA,CUUUUG,CUUUUU,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCU,UUUUGU,UUUUUA,UUUUUC,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| NOVA2 |
9 |
1434 |
0.023809524 |
0.0072299476 |
1.719482 |
AGCACC,UUUUUU |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| SF1 |
8 |
1294 |
0.021428571 |
0.0065245869 |
1.715577 |
ACUAGC,ACUUAU,AGUAAG,AUACUA,CUAACA,UAACAA,UAUACU,UGCUAA |
ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| ELAVL2 |
67 |
9826 |
0.161904762 |
0.0495112858 |
1.709316 |
AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUUA,AUUUUC,AUUUUU,CAUUUU,CUUUUA,CUUUUU,GUUUUU,UAAUUU,UACUUU,UAUACU,UAUAUA,UAUAUU,UAUUUA,UAUUUU,UCAUUU,UCUUUU,UUAAUU,UUACUU,UUAUUU,UUCAUU,UUGAUU,UUUAAU,UUUACU,UUUAUU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL1 |
19 |
3309 |
0.047619048 |
0.0166767432 |
1.513701 |
UAUUUA,UAUUUU,UUAUUU,UUGAUU,UUUAUU,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| PUM1 |
12 |
2172 |
0.030952381 |
0.0109482064 |
1.499356 |
AAUUGU,AGAUAA,AUUGUA,CAGAAU,CUUGUA,UAAAUA,UAUAUA,UGUAAA,UUGUAC,UUUAAU |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| RBM28 |
1 |
340 |
0.004761905 |
0.0017180572 |
1.470761 |
UGUAGG |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| QKI |
4 |
904 |
0.011904762 |
0.0045596534 |
1.384543 |
ACUCAU,ACUUAU,CUAACA,UACUCA |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPD |
7 |
1488 |
0.019047619 |
0.0075020153 |
1.344261 |
UAUUUA,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| FUS |
8 |
1711 |
0.021428571 |
0.0086255542 |
1.312847 |
UUUUUU |
AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| PABPC5 |
2 |
596 |
0.007142857 |
0.0030078597 |
1.247764 |
AGAAAU,GAAAUU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| DAZAP1 |
13 |
2878 |
0.033333333 |
0.0145052398 |
1.200391 |
AGGUUA,AGUAAG,AGUAUA,UAGGUU,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ZCRB1 |
2 |
666 |
0.007142857 |
0.0033605401 |
1.087808 |
AAUUAA,AUUUAA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CELF5 |
2 |
669 |
0.007142857 |
0.0033756550 |
1.081334 |
GUGUUG,GUGUUU |
GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| AGO2 |
2 |
677 |
0.007142857 |
0.0034159613 |
1.064210 |
UAAAGU |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |