circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000198876:-:9:34106433:34107565
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198876:-:9:34106433:34107565 | ENSG00000198876 | ENST00000361264 | - | 9 | 34106434 | 34107565 | 268 | CUAUUUGUCGUAGAUGUCCAGACAAGCCAGAUCACCAAGAUCCCCAUUCUGAAAGACCGGGAGCCUGGAGGUGUGACCCAGCAGGGCUGUGGUAUCCAUGCCAUCGAGCUGAAUCCUUCUAGAACACUGCUAGCCACUGGAGGAGACAACCCCAACAGUCUUGCCAUCUAUCGACUACCUACGCUGGAUCCUGUGUGUGUAGGAGAUGAUGGACACAAGGACUGGAUCUUUUCCAUCGCAUGGAUCAGCGACACUAUGGCAGUGUCUGCUAUUUGUCGUAGAUGUCCAGACAAGCCAGAUCACCAAGAUCCCCAUUCU | circ |
| ENSG00000198876:-:9:34106433:34107565 | ENSG00000198876 | ENST00000361264 | - | 9 | 34106434 | 34107565 | 22 | GGCAGUGUCUGCUAUUUGUCGU | bsj |
| ENSG00000198876:-:9:34106433:34107565 | ENSG00000198876 | ENST00000361264 | - | 9 | 34107556 | 34107765 | 210 | AUCUCUAAUAUUGUGUUAUAUUGACUUAUUUCAUGUUUCAUGUCAUUAGUAGUCCUGACCAUGCCUUUAAUAAUUCCUAGGGUCUAACAUGUAGUAGUUGUUAAUUAAAUGUUGAUGAACAGUUGAACAAAAGGAUGUUUAUACCCUGUAUUGGCCACGUUAAAUUUAUGUUCAGCUUCUGUAUCCAUUUUUUCUUGUAGCUAUUUGUCG | ie_up |
| ENSG00000198876:-:9:34106433:34107565 | ENSG00000198876 | ENST00000361264 | - | 9 | 34106234 | 34106443 | 210 | GCAGUGUCUGGUAAAGUUGCCCUUUUAUAGGGAGCUUUUGAUGUGGCAGGGAUUGGGGGCUAGAGAUUGAAAAGGGAUAGUAGUGUGAUAGGAAACAGCCCAGCCGGGCUCAGUGACUCACGCCUAUAAUCCCAGCACUUUGGGAGGCUGAGGUGGGUGGAUCACUUGAGCCCAAGAAUUUAAAUUAGUUGAGUGUGGUACUGCAUACUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
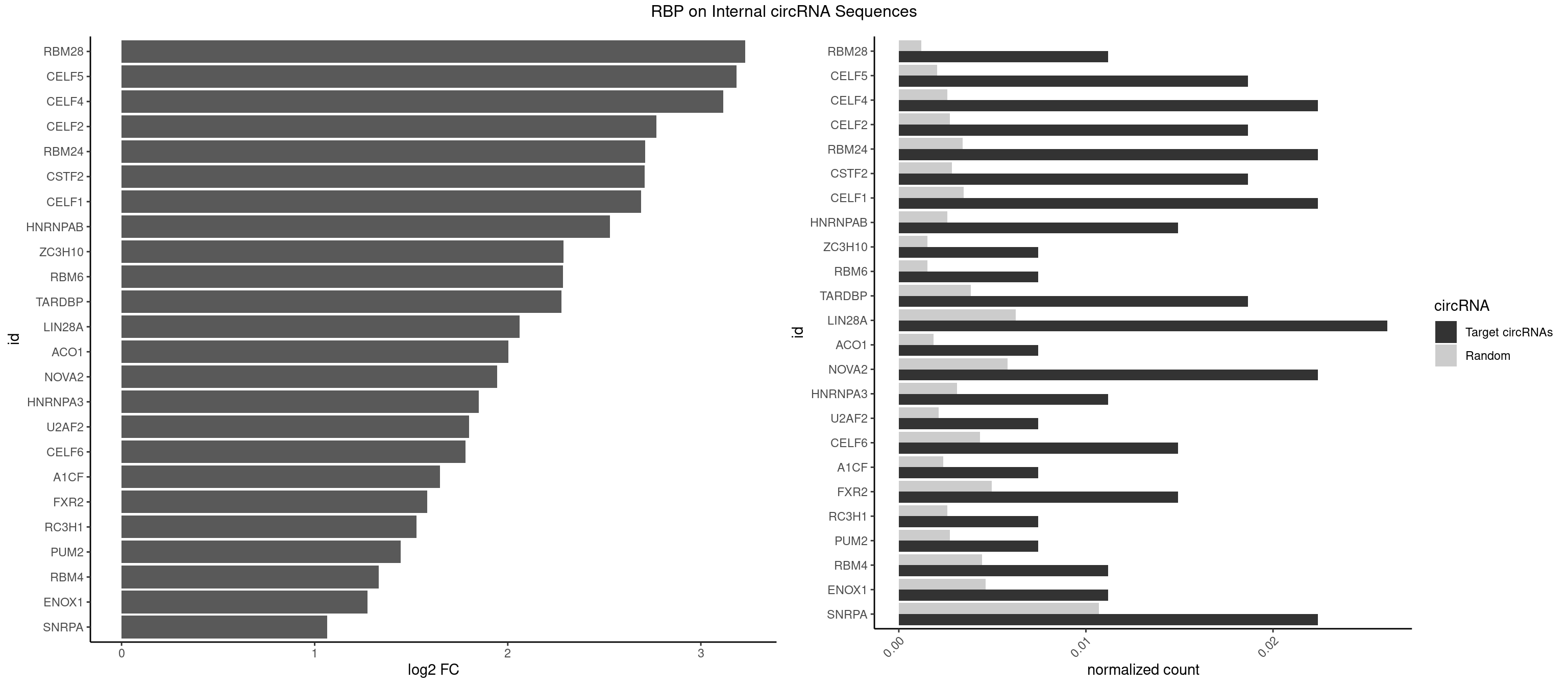
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM28 | 2 | 822 | 0.011194030 | 0.001192695 | 3.230433 | GUGUAG,UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| CELF5 | 4 | 1415 | 0.018656716 | 0.002052073 | 3.184541 | GUGUGU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 5 | 1782 | 0.022388060 | 0.002583931 | 3.115090 | GGUGUG,GUGUGU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF2 | 4 | 1886 | 0.018656716 | 0.002734648 | 2.770268 | GUGUGU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM24 | 5 | 2357 | 0.022388060 | 0.003417223 | 2.711833 | GUGUGA,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CSTF2 | 4 | 1967 | 0.018656716 | 0.002852033 | 2.709632 | GUGUGU,UGUGUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF1 | 5 | 2391 | 0.022388060 | 0.003466496 | 2.691180 | GUGUGU,UGUCUG,UGUGUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPAB | 3 | 1782 | 0.014925373 | 0.002583931 | 2.530128 | AAAGAC,AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ZC3H10 | 1 | 1053 | 0.007462687 | 0.001527461 | 2.288560 | CAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBM6 | 1 | 1054 | 0.007462687 | 0.001528910 | 2.287191 | UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| TARDBP | 4 | 2654 | 0.018656716 | 0.003847636 | 2.277651 | GUGUGU,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| LIN28A | 6 | 4315 | 0.026119403 | 0.006254764 | 2.062095 | AGGAGA,GGAGAU,GGAGGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| ACO1 | 1 | 1283 | 0.007462687 | 0.001860778 | 2.003789 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| NOVA2 | 5 | 4013 | 0.022388060 | 0.005817105 | 1.944356 | AGAUCA,AUCACC,GAGACA,GAUCAC | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| HNRNPA3 | 2 | 2140 | 0.011194030 | 0.003102746 | 1.851112 | CAAGGA,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| U2AF2 | 1 | 1477 | 0.007462687 | 0.002141923 | 1.800788 | UUUUCC | UUUUCC,UUUUUC,UUUUUU |
| CELF6 | 3 | 2997 | 0.014925373 | 0.004344713 | 1.780434 | UGUGGU,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| A1CF | 1 | 1642 | 0.007462687 | 0.002381042 | 1.648102 | GAUCAG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| FXR2 | 3 | 3434 | 0.014925373 | 0.004978016 | 1.584124 | AGACAA,GACAAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| RC3H1 | 1 | 1786 | 0.007462687 | 0.002589727 | 1.526895 | CCUUCU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PUM2 | 1 | 1890 | 0.007462687 | 0.002740445 | 1.445285 | GUAGAU | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| RBM4 | 2 | 3065 | 0.011194030 | 0.004443259 | 1.333039 | CCUUCU,UCCUUC | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ENOX1 | 2 | 3195 | 0.011194030 | 0.004631656 | 1.273130 | CAGACA,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| SNRPA | 5 | 7380 | 0.022388060 | 0.010696574 | 1.065581 | AGGAGA,CUGCUA,GGAGAU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
RBP on BSJ (Exon Only)
Plot

Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| YBX1 | 1 | 19 | 0.09090909 | 0.001309929 | 6.116864 | GUCUGC | AACCAC,ACACCA,ACAUCU,ACCACC,CACCAC,CAGCAA,CCACCA,CCAGCA,CCUGCG,CUGCGG,GAUCUG,GCCUGC,UCCAGC,UGCGGU |
| MBNL1 | 2 | 36 | 0.13636364 | 0.002423369 | 5.814301 | CUGCUA,UCUGCU | AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUG,CCUGCU,CGCUGC,CGCUGU,CUGCCG,CUGCCU,CUGCGG,CUGCUC,CUGCUU,CUUGCU,GCUUGC,GCUUGU,GGCUUU,GUGCUG,UGCGGC,UGCUGC,UGCUGU,UGCUUU,UUGCCU,UUGCUC,UUGCUG,UUGCUU,UUGUGC |
| SNRPA | 1 | 39 | 0.09090909 | 0.002619859 | 5.116864 | CUGCUA | AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,GAGCAG,GAUACC,GCAGUA,GGAGAU,GGGUAU,GUAGGC,GUAGUG,UAUGCU,UCCUGC,UGCACG,UUCCUG,UUGCAC |
| SRSF2 | 1 | 479 | 0.09090909 | 0.031438302 | 1.531901 | CUGCUA | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
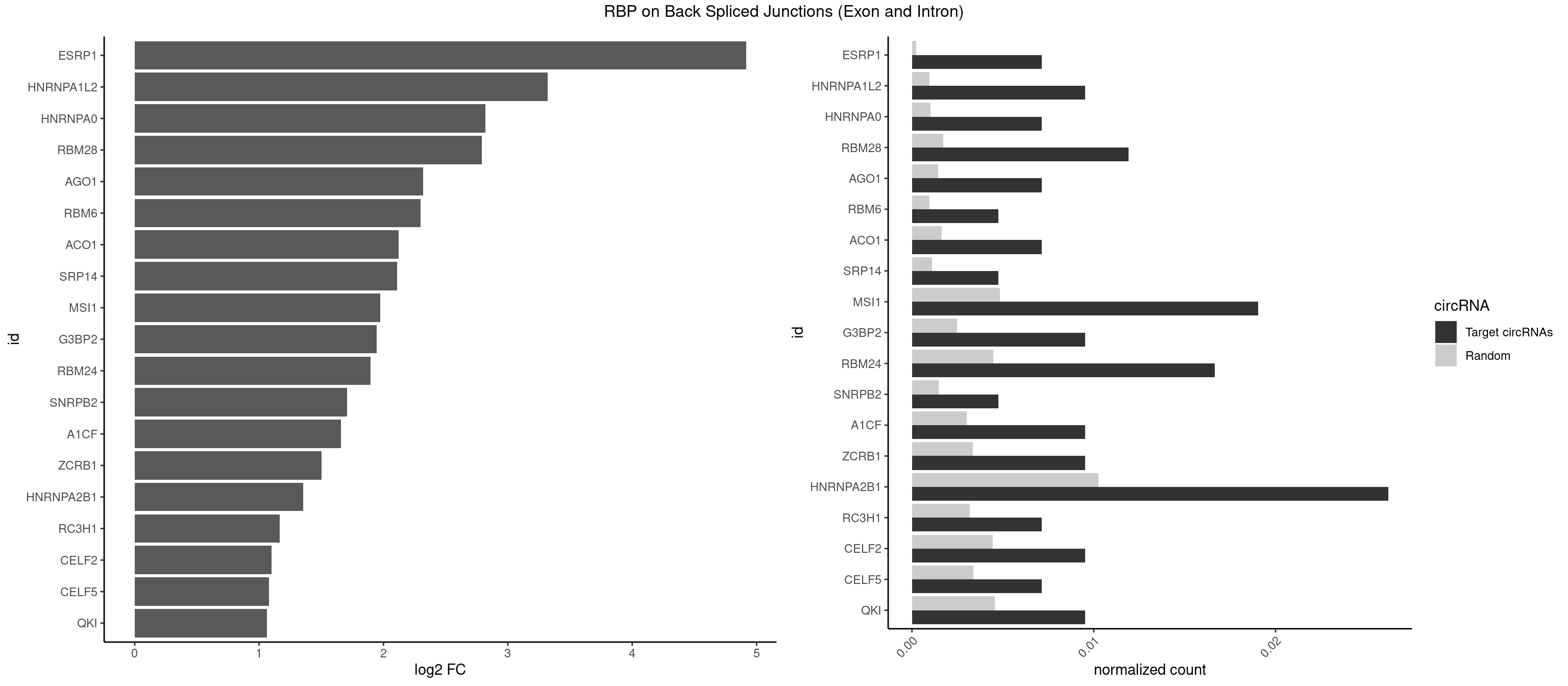
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 2 | 46 | 0.007142857 | 0.0002367997 | 4.914762 | AGGGAU | AGGGAU |
| HNRNPA1L2 | 3 | 188 | 0.009523810 | 0.0009522370 | 3.322146 | AUAGGG,UAGGGA,UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| RBM28 | 4 | 340 | 0.011904762 | 0.0017180572 | 2.792689 | AGUAGU,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | GUAGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBM6 | 1 | 191 | 0.004761905 | 0.0009673519 | 2.299426 | UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CCUGUA | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| MSI1 | 7 | 961 | 0.019047619 | 0.0048468360 | 1.974496 | AGGUGG,UAGGAA,UAGUAG,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUG,GGAUAG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM24 | 6 | 888 | 0.016666667 | 0.0044790407 | 1.895704 | AGUGUG,GAGUGU,GUGUGA,GUGUGG,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AAUUAA,AUUUAA,GACUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPA2B1 | 10 | 2033 | 0.026190476 | 0.0102478839 | 1.353716 | AAUUUA,AUAGGG,AUUUAA,CAAGAA,CCAAGA,UAGGGA,UAGGGU,UUUAUA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CUUCUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUUGUU,UAUGUU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUGG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | ACUUAU,CUAACA,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.