circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000113558:-:5:134157706:134167243
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000113558:-:5:134157706:134167243 | ENSG00000113558 | MSTRG.26934.1 | - | 5 | 134157707 | 134167243 | 421 | AUUUGGGAAUGGAUGAUGAAGGAGAUGAUGACCCAGUUCCUCUACCAAAUGUGAAUGCAGCAAUAUUAAAAAAGGUCAUUCAGUGGUGCACCCACCACAAGGAUGACCCUCCUCCUCCUGAAGAUGAUGAGAACAAAGAAAAGCGAACAGAUGAUAUCCCUGUUUGGGACCAAGAAUUCCUGAAAGUUGACCAAGGAACACUUUUUGAACUCAUUCUGGCUGCAAACUACUUAGACAUCAAAGGUUUGCUUGAUGUUACAUGCAAGACUGUUGCCAAUAUGAUCAAGGGGAAAACUCCUGAGGAGAUUCGCAAGACCUUCAAUAUCAAAAAUGACUUUACUGAAGAGGAGGAAGCCCAGGUACGCAAAGAGAACCAGUGGUGUGAAGAGAAGUGAAAUGUUGUGCCUGACACUGUAACACUAUUUGGGAAUGGAUGAUGAAGGAGAUGAUGACCCAGUUCCUCUACCAAAU | circ |
| ENSG00000113558:-:5:134157706:134167243 | ENSG00000113558 | MSTRG.26934.1 | - | 5 | 134157707 | 134167243 | 22 | ACUGUAACACUAUUUGGGAAUG | bsj |
| ENSG00000113558:-:5:134157706:134167243 | ENSG00000113558 | MSTRG.26934.1 | - | 5 | 134167234 | 134167443 | 210 | CCAGGAAACAAGUCUACAUGUUUAGUGCAAGACACAGCCAUCCAUUUUUUUUCCCAAUAUGUUUAAUCUGUGGUUGGUUGAAUCCACAGAUGUAGAACUCAUGGAUAUGGGGGCUGACUCUUAUGUUUUGAGACAUAGCCUGGUUGGUAUAAGUAUUAUAAUGGAAUUAGGAAAUAGAAACUUUCUUCUGGUUUUCUUAGAUUUGGGAAU | ie_up |
| ENSG00000113558:-:5:134157706:134167243 | ENSG00000113558 | MSTRG.26934.1 | - | 5 | 134157507 | 134157716 | 210 | CUGUAACACUGUAAGGAUUGUUCCAAAUACUAGUUGCACUGCUCUGUUUAUAAUUGUUAAUAUUAGACAAACAGUAGACAAAUGCAGCAGCAAGUCAAUUGUAUUAGCAGAAUAUUGUCCUCAUUGCAUGUGUAGUUUGAGCACAGAUCCCAAACCUUACGGCCAAGUUUCUUCUAGUAUGAUGGAAAGUUUCUUUUUUCUUUGCUCUGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
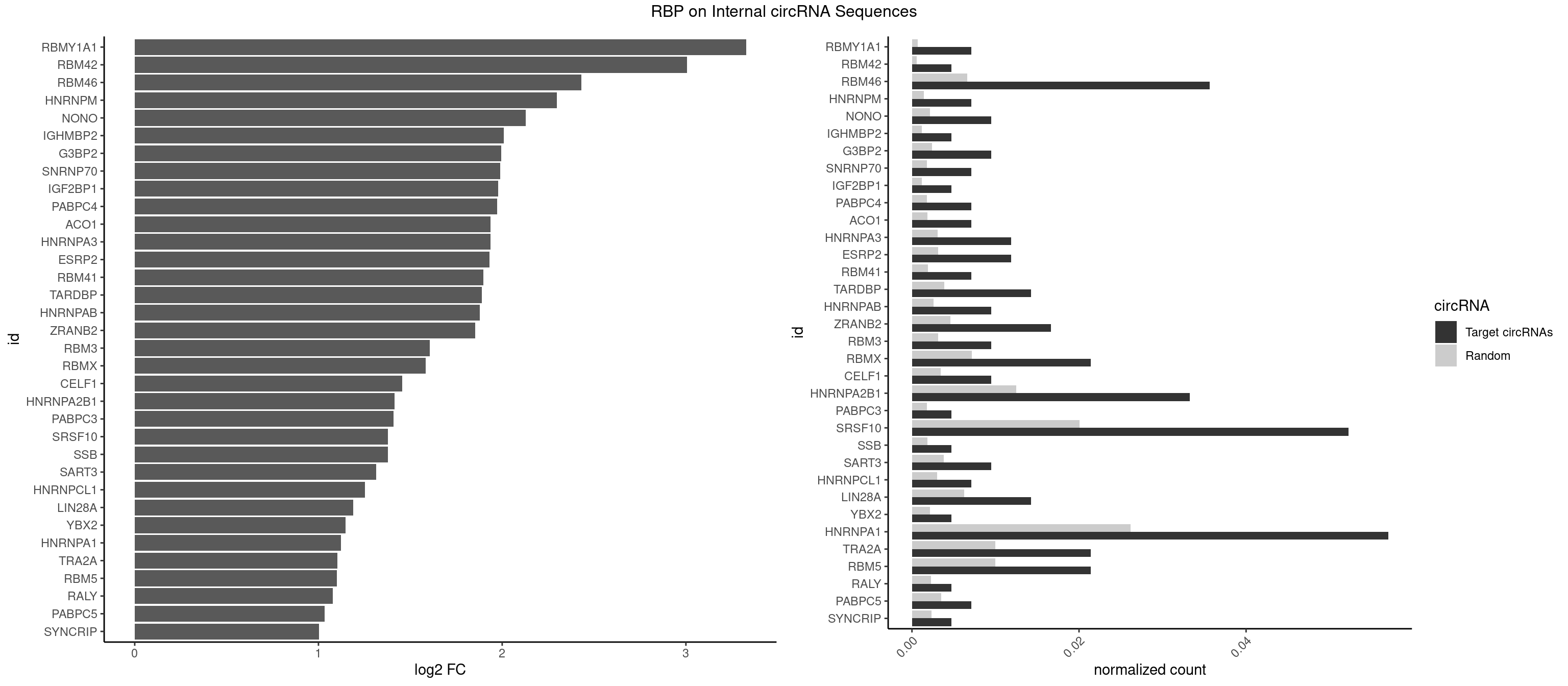
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 2 | 489 | 0.007125891 | 0.0007101099 | 3.326956 | CAAGAC | ACAAGA,CAAGAC |
| RBM42 | 1 | 407 | 0.004750594 | 0.0005912752 | 3.006206 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBM46 | 14 | 4554 | 0.035629454 | 0.0066011240 | 2.432287 | AUCAAA,AUCAAG,AUGAAG,AUGAUA,AUGAUG,GAUCAA,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPM | 2 | 999 | 0.007125891 | 0.0014492040 | 2.297810 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| NONO | 3 | 1498 | 0.009501188 | 0.0021723567 | 2.128847 | AGAGGA,AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| IGHMBP2 | 1 | 813 | 0.004750594 | 0.0011796520 | 2.009747 | AAAAAA | AAAAAA |
| G3BP2 | 3 | 1644 | 0.009501188 | 0.0023839405 | 1.994760 | AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SNRNP70 | 2 | 1237 | 0.007125891 | 0.0017941145 | 1.989798 | AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| IGF2BP1 | 1 | 831 | 0.004750594 | 0.0012057377 | 1.978192 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PABPC4 | 2 | 1251 | 0.007125891 | 0.0018144033 | 1.973575 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| ACO1 | 2 | 1283 | 0.007125891 | 0.0018607779 | 1.937165 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPA3 | 4 | 2140 | 0.011876485 | 0.0031027457 | 1.936491 | AAGGAG,CAAGGA,CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 4 | 2150 | 0.011876485 | 0.0031172377 | 1.929768 | GGGAAA,GGGAAU,GGGGAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM41 | 2 | 1318 | 0.007125891 | 0.0019115000 | 1.898365 | UACAUG,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| TARDBP | 5 | 2654 | 0.014251781 | 0.0038476365 | 1.889098 | GAAUGG,GUGAAU,GUUGUG,UGAAUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| HNRNPAB | 3 | 1782 | 0.009501188 | 0.0025839306 | 1.878541 | ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ZRANB2 | 6 | 3173 | 0.016627078 | 0.0045997733 | 1.853900 | AAAGGU,AGGUAC,AGGUUU,GUGGUG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| RBM3 | 3 | 2152 | 0.009501188 | 0.0031201361 | 1.606499 | AAAACU,AAACUA,AAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBMX | 8 | 4925 | 0.021377672 | 0.0071387787 | 1.582356 | AAGGAA,ACCAAA,AGGAAG,AUCAAA,GAAGGA,GUAACA,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| CELF1 | 3 | 2391 | 0.009501188 | 0.0034664959 | 1.454630 | GUUGUG,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPA2B1 | 13 | 8607 | 0.033254157 | 0.0124747476 | 1.414524 | AAGGAA,AAGGAG,AAGGGG,AGGAAC,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,UAGACA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| PABPC3 | 1 | 1234 | 0.004750594 | 0.0017897669 | 1.408336 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SRSF10 | 21 | 13860 | 0.052256532 | 0.0200874160 | 1.379319 | AAAGAA,AAAGAG,AAGAAA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGG,ACAAAG,AGAGAA,AGAGGA,CAAAGA,GAGAAC,GAGAAG,GAGGAA,GAGGAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SSB | 1 | 1260 | 0.004750594 | 0.0018274462 | 1.378279 | CUGUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SART3 | 3 | 2634 | 0.009501188 | 0.0038186524 | 1.315044 | AAAAAA,AGAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| HNRNPCL1 | 2 | 2062 | 0.007125891 | 0.0029897078 | 1.253066 | CUUUUU,UUUUUG | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| LIN28A | 5 | 4315 | 0.014251781 | 0.0062547643 | 1.188115 | AGGAGA,GGAGAU,GGAGGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| YBX2 | 1 | 1480 | 0.004750594 | 0.0021462711 | 1.146276 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| HNRNPA1 | 23 | 18070 | 0.057007126 | 0.0261885646 | 1.122205 | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AGAGGA,AGGAAG,AGUGAA,AGUGGU,CAAAGA,CAAGGA,CCAAGG,GAGGAA,GAGGAG,GGAGGA,GUACGC,GUGGUG,UAGACA,UGGUGC | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| TRA2A | 8 | 6871 | 0.021377672 | 0.0099589296 | 1.102042 | AAAGAA,AAGAAA,AAGAGG,AGAGGA,AGGAAG,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RBM5 | 8 | 6879 | 0.021377672 | 0.0099705232 | 1.100364 | AAAAAA,AAGGAA,AAGGAG,AAGGGG,CAAGGA,CAAGGG,GAAGGA | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| RALY | 1 | 1553 | 0.004750594 | 0.0022520629 | 1.076861 | UUUUUG | UUUUUC,UUUUUG,UUUUUU |
| PABPC5 | 2 | 2400 | 0.007125891 | 0.0034795387 | 1.034174 | AGAAAA,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SYNCRIP | 1 | 1634 | 0.004750594 | 0.0023694485 | 1.003557 | AAAAAA | AAAAAA,UUUUUU |
RBP on BSJ (Exon Only)
Plot
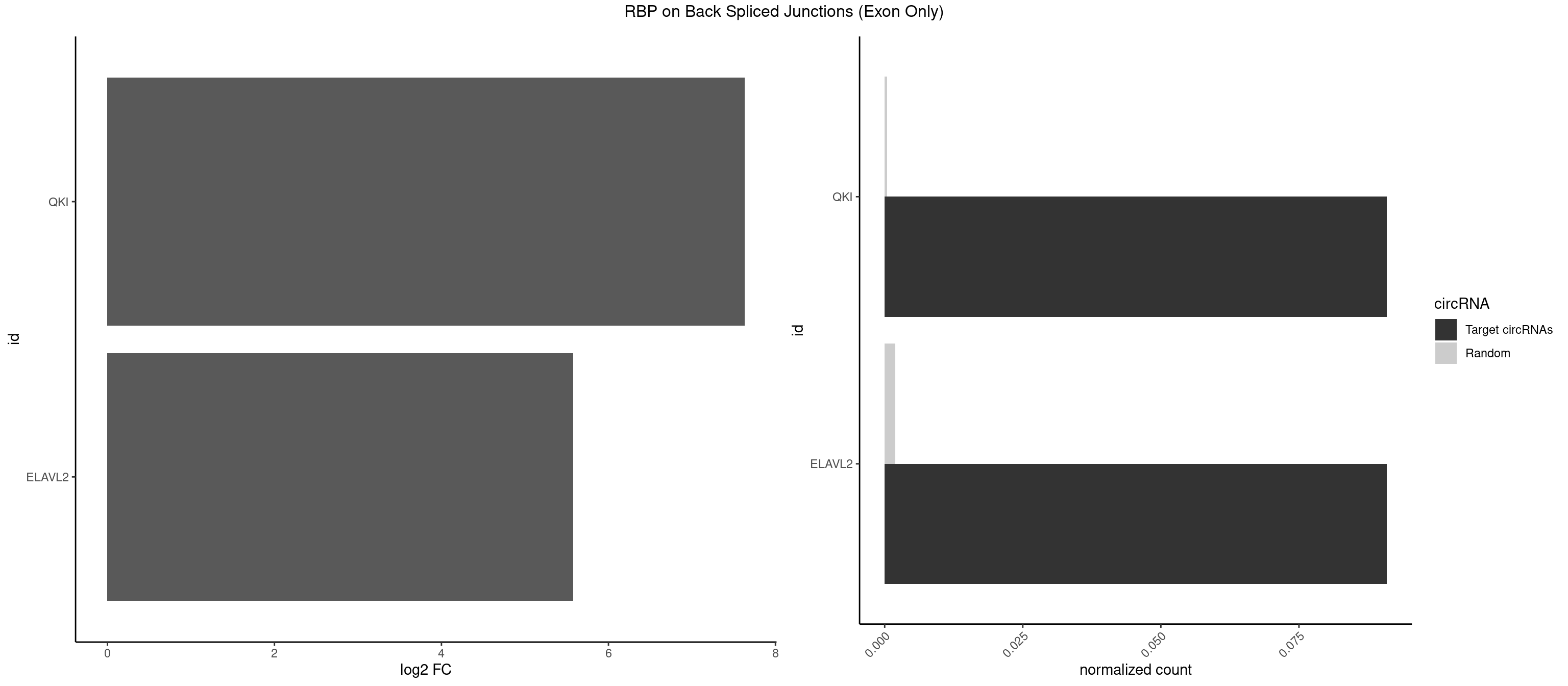
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| QKI | 1 | 6 | 0.09090909 | 0.0004584752 | 7.631437 | ACACUA | AUCUAA,UAAUCA,UCUAAU |
| ELAVL2 | 1 | 28 | 0.09090909 | 0.0018993974 | 5.580811 | UAUUUG | AAUUUA,AAUUUU,AUAUUU,AUUUAA,AUUUUG,CUUUCU,GAUUUU,GUAUUG,GUUUUU,UACUUU,UAGUUA,UAUAUA,UAUAUU,UAUGUU,UAUUGA,UAUUUU,UGUAUU,UUAAGU,UUACUU,UUAGUU,UUAUUG,UUUACU,UUUAGU,UUUCUU,UUUUAG |
RBP on BSJ (Exon and Intron)
Plot
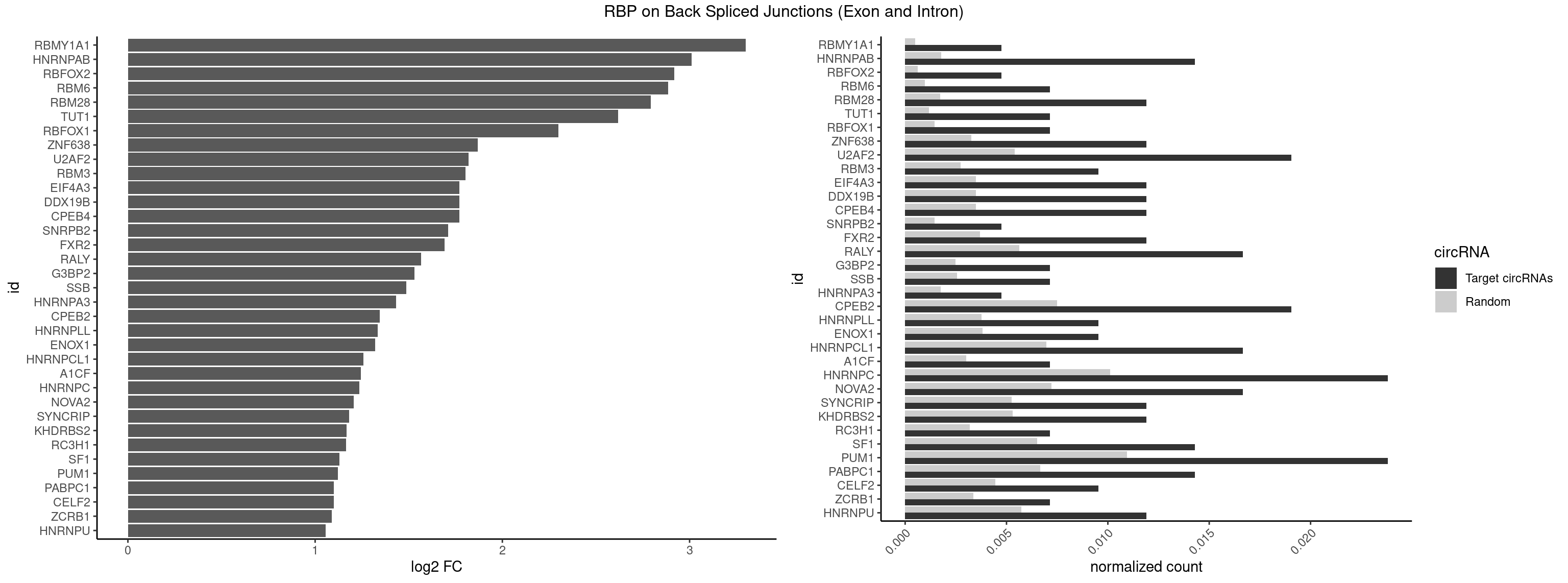
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| HNRNPAB | 5 | 351 | 0.014285714 | 0.0017734784 | 3.009919 | AAGACA,AGACAA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBM28 | 4 | 340 | 0.011904762 | 0.0017180572 | 2.792689 | AGUAGA,GUGUAG,UGUAGA,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| ZNF638 | 4 | 646 | 0.011904762 | 0.0032597743 | 1.868695 | GGUUGG,GUUGGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| U2AF2 | 7 | 1071 | 0.019047619 | 0.0054010480 | 1.818299 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAUACU,AUACUA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CPEB4 | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| DDX19B | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| EIF4A3 | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | AUUGCA | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| FXR2 | 4 | 730 | 0.011904762 | 0.0036829907 | 1.692589 | AGACAA,GACAAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| RALY | 6 | 1117 | 0.016666667 | 0.0056328094 | 1.565039 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUU,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| CPEB2 | 7 | 1487 | 0.019047619 | 0.0074969770 | 1.345230 | AUUUUU,CAUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPLL | 3 | 748 | 0.009523810 | 0.0037736800 | 1.335567 | ACAAAC,CAAACA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AAGACA,UAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPCL1 | 6 | 1381 | 0.016666667 | 0.0069629182 | 1.259202 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUAAUU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPC | 9 | 2006 | 0.023809524 | 0.0101118501 | 1.235492 | AUUUUU,CUUUUU,GGAUAU,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| NOVA2 | 6 | 1434 | 0.016666667 | 0.0072299476 | 1.204908 | AGACAU,GAGACA,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| SYNCRIP | 4 | 1041 | 0.011904762 | 0.0052498992 | 1.181177 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CUUCUG,UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | AUACUA,CACAGA,GCUGAC,UACUAG | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| PUM1 | 9 | 2172 | 0.023809524 | 0.0109482064 | 1.120844 | AAUAUU,AAUUGU,AUUGUA,CAGAAU,UAAUAU,UGUAGA,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| PABPC1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | ACAAAC,ACAAAU,CAAACA,CAAACC,CAAAUA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | AUGUGU,UAUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAAUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPU | 4 | 1136 | 0.011904762 | 0.0057285369 | 1.055300 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.