circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000139746:-:13:79319534:79378907
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000139746:-:13:79319534:79378907 | ENSG00000139746 | MSTRG.8946.3 | - | 13 | 79319535 | 79378907 | 3967 | CUGUGAUGCAGAUCCAUCCGCCCUAGCAAAAUAUGUUCUGGCUUUGGUAAAGAAAGACAAAAGUGAAAAAGAGUUAAAGGCAUUAUGUAUUGAUCAGCUGGAUGUAUUUCUUCAGAAAGAGACACAGAUAUUUGUGGAAAAACUUUUUGAUGCUGUGAAUACAAAGAGUUACCUACCUCCUCCAGAGCAGCCAUCAUCAGGAAGCCUGAAGGUAGAAUUUUUUCCACACCAAGAAAAAGAUAUAAAGAAGGAAGAGAUCACUAAGGAGGAAGAGCGAGAGAAGAAGUUUUCUAGAAGGCUAAAUCACAGUCCUCCCCAGUCAAGCUCCCGAUACAGGGAAAAUAGAAGCCGUGAUGAGAGGAAAAAAGAUGAUCGUUCUCGCAAAAGAGAUUAUGAUCGAAACCCUCCUCGAAGAGAUUCAUACAGAGACCGGUACAAUAGAAGACGAGGGCGGAGUCGCAGUUAUAGCAGGAGUCGAAGUCGAAGUUGGAGUAAAGAGAGGCUUCGUGAGAGGGACAGAGAUAGAAGCAGGACUAGAAGCAGAAGCAGAACACGAAGCAGGGAAAGGGAUCUGGUAAAACCUAAAUAUGACCUGGAUAGAACAGAUCCAUUAGAAAAUAAUUAUACUCCAGUCUCUUCGGUACCUAGUAUUUCAUCUGGCCACUACCCUGUACCUACUUUGAGCAGCACUAUUACAGUAAUUGCUCCUACUCAUCAUGGAAACAACACUACCGAAAGUUGGUCUGAAUUUCAUGAAGACCAAGUGGACCAUAACUCUUACGUAAGACCACCCAUGCCAAAGAAACGGUGUAGAGACUAUGAUGAAAAGGGUUUUUGUAUGAGAGGAGACAUGUGUCCUUUUGAUCAUGGAAGUGAUCCAGUAGUUGUAGAAGAUGUGAAUCUUCCUGGUAUGCUGCCUUUCCCAGCACAGCCUCCUGUUGUUGAAGGACCACCUCCUCCUGGACUCCCCCCACCUCCACCAAUUCUUACACCCCCACCUGUGAAUCUCAGGCCCCCAGUACCACCGCCAGGUCCAUUGCCACCCAGUCUCCCACCUGUUACAGGACCACCACCUCCACUUCCUCCUUUGCAGCCAUCUGGCAUGGAUGCUCCUCCAAACUCUGCAACCAGUUCUGUUCCUACUGUAGUAACAACUGGCAUUCAUCACCAGCCUCCUCCUGCUCCACCCUCUCUUUUUACUGCAGAUACAUAUGACACAGAUGGCUACAAUCCUGAAGCCCCAAGCAUAACAAACACUUCCAGACCUAUGUAUAGACACAGAGUGCAUGCACAAAGGCCCAACUUGAUAGGACUAACAUCAGGGGAUAUGGAUUUGCCACCCAGAGAAAAGCCUCCCAAUAAAAGCAGUAUGAGGAUAGUAGUGGACUCAGAAUCAAGGAAAAGAACCAUUGGUUCUGGAGAGCCUGGAGUUCCUACAAAGAAGACUUGGUUUGAUAAACCAAAUUUUAAUAGAACAAACAGCCCAGGCUUUCAGAAGAAGGUUCAAUUUGGAAAUGAAAAUACCAAGCUUGAACUUAGAAAAGUUCCUCCAGAAUUAAAUAAUAUCAGCAAACUUAAUGAACAUUUUAGUCGAUUUGGAACCUUGGUUAACUUACAGGUUGCUUAUAAUGGUGAUCCUGAAGGUGCCCUAAUCCAAUUUGCAACAUACGAAGAAGCAAAGAAAGCAAUAUCAAGUACGGAAGCAGUAUUAAACAAUCGCUUUAUUAAGGUUUAUUGGCACAGAGAAGGAAGCACCCAACAGUUACAAACUACUUCUCCAAAGGUAAUGCAGCCUUUAGUCCAGCAGCCCAUUUUGCCUGUUGUGAAGCAGUCAGUCAAAGAGCGGCUGGGUCCAGUACCUUCAAGUACUAUUGAACCUGCAGAAGCCCAGAGUGCCUCUUCAGACCUUCCUCAGGUGUUGUCUACAUCUACUGGCCUAACAAAAACAGUGUAUAAUCCAGCUGCUUUGAAGGCUGCACAGAAAACCUUACUUGUUUCCACCUCUGCAGUUGAUAAUAAUGAAGCACAGAAAAAAAAACAGGAGGCAUUGAAACUUCAGCAGGAUGUAAGGAAAAGGAAACAAGAAAUUUUAGAAAAGCACAUUGAAACACAGAAGAUGUUAAUUUCAAAACUGGAGAAAAACAAAACAAUGAAGUCUGAAGAUAAAGCAGAAAUAAUGAAAACUUUAGAGGUUUUGACAAAAAAUAUUACCAAGUUGAAAGAUGAGGUCAAAGCUGCUUCUCCUGGACGCUGUCUUCCAAAAAGUAUAAAAACCAAGACUCAGAUGCAGAAGGAAUUACUUGACACAGAACUGGAUUUAUAUAAGAAGAUGCAGGCUGGAGAAGAAGUCACUGAACUUAGGAGAAAGUAUACAGAAUUACAGCUGGAAGCUGCCAAACGAGGGAUUCUUUCAUCUGGUCGGGGCAGAGGAAUUCAUUCAAGAGGUCGAGGUGCAGUUCAUGGCCGAGGCAGGGGGCGAGGGCGAGGGCGAGGUGUGCCUGGUCAUGCUGUGGUGGAUCACCGUCCCAGGGCAUUGGAGAUUUCUGCAUUUACGGAGAGCGAUAGAGAAGAUCUUCUUCCUCAUUUUGCGCAAUAUGGUGAAAUUGAAGAUUGUCAGAUUGAUGAUUCCUCACUUCAUGCAGUAAUUACAUUCAAGACAAGAGCAGAAGCUGAAGCAGCUGCAGUUCAUGGAGCUCGUUUCAAAGGGCAAGAUCUAAAACUGGCAUGGAAUAAACCAGUAACUAAUAUUUCAGCUGUUGAAACAGAAGAAGUUGAGCCUGAUGAAGAAGAAUUUCAGGAAGAGUCUUUGGUGGAUGACUCAUUACUUCAAGAUGAUGAUGAAGAAGAAGAGGACAAUGAAUCUCGUUCUUGGAGAAGAUGAUUUGACUGAUCAUUGAUCUGCAUAUGCUAGAACUCUACCUGUGUUUCAUUAGUAUUAUCUAAUGUACUUUUACAUAUUUGUAAAAACAAUUUUUGGUAAAAUGUGAUGAAGAUGGAUUUCACAAAUAGACAAAAAAGAAGAAAACUACCUUCUGAUCUUGUAUUUUGAAAGAUUGAUGUUUGCAUUUUACUUCAGUAAACAAUUGCUAAAGACAUCACACUAGAAACAUAUGCAAUGUUUUUAUUACAUACUUCUACUGGACAUCACAGAAUUCUUUGGGUUCUUUGUAAUUUAAUGAAUAGGUCUGAAAACUUAUGACCAAUACUUGUUAUAACUUAGAGGACUUUGUUUUAUUCCAAAUAAGGAAUGAAUUUGCAUUUAAAAUCUUAAUGAAUGUUUUCAAAACUGAAUAGAUAACAUAGUACUCUAACUAAAGUCUCCAAGUUAUGUAUUAUAAUAUUACAUAGUAGUAUGCUUAGGCUUUACUAUGUAUUAGCCUUUUGUUGGACUGUGUAUGUAUUUUACCAUAUGGGUUUUAAUGAUAAUGGUGUAUGACUGCUUUACAUGAGUCCUUAUGCAUCCAGAUGUUAUAAUAAAGUGGAAUGGUCUCUUUAAAAAAAAAAAAGGAAAGAAAAGAGAAAAGCAAUGACAAAAAAAAAAAAAAAAAAAAAGCCAAGACAAUGUUCCUUGAUUUAAAAAAAAAAAGAAAAGAAAAAAGAAAAGAAAAAAAUUAAAAUAGACCAUUCCAGAUGGUGAUCUACCCUUCCCCCCAAUUAUCACAAAAGGAGCUAUAAUCACUAACUUAUUCUUAAUAAUAAUGGUUACUGGUACCCUUACAAUAAUGUACAAUCCAAUGUUUAAAAAUUAUACCACUUGAGUUUGGUUUGAUCCUAUAAAUUUUCAACAAAUGUCUUAAAAUUUAAAAGCAGGCUGAUUAGUUUGGAACCAGACUGCAAGUAGAGCUAACAUUUGGUGAAUAAGAAAACAUCACAUUACAUUUUGGAUGUAUGAAAGAAAACUUGACCAUUUGAAGAUAUAUGAAUAAAGAAAUGUACUAUAGAUACUACUUUAGGAAAACACUGUUUGCUGUGAUGCAGAUCCAUCCGCCCUAGCAAAAUAUGUUCUGGCUUUGGUAA | circ |
| ENSG00000139746:-:13:79319534:79378907 | ENSG00000139746 | MSTRG.8946.3 | - | 13 | 79319535 | 79378907 | 22 | AACACUGUUUGCUGUGAUGCAG | bsj |
| ENSG00000139746:-:13:79319534:79378907 | ENSG00000139746 | MSTRG.8946.3 | - | 13 | 79378898 | 79379107 | 210 | AAGAAAUAAAAUGGCUACACAUAGCAUUAAGGAGUGUUAGUAUAAAGAGAACUUAAAUUUCUUCAAGGUCCUCUGUGACCCGGGUGCUUUUUACAUGUAUUAACUCACUUAACUAUUAUUCUCACUAAUUCAGUUUGUAACUGGAAUUUGUAGAAUGUGCAGUUGGCUAAAUUUUCUUCAACAUUGGUUUUUUUCUUUAGCUGUGAUGCA | ie_up |
| ENSG00000139746:-:13:79319534:79378907 | ENSG00000139746 | MSTRG.8946.3 | - | 13 | 79319335 | 79319544 | 210 | ACACUGUUUGGUAAGUGUUCCAGUCUGAUAUUUUAAGUGUGUACUUCCUAUACUUGUAUAAAAAUCUUUGAUGUUUGCAACAAUACUGUGAUUUUUUUAAGUGGAUUUCUUUUGCAAGUGAACAUGAUCAUUGAAAGUUGAUAUGGCAGUUUCCAGGAAAUAAAUGCAUUCUUUGUAGAAUGGGGGUGGGAGAUUACAUAUUUAUAUUUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
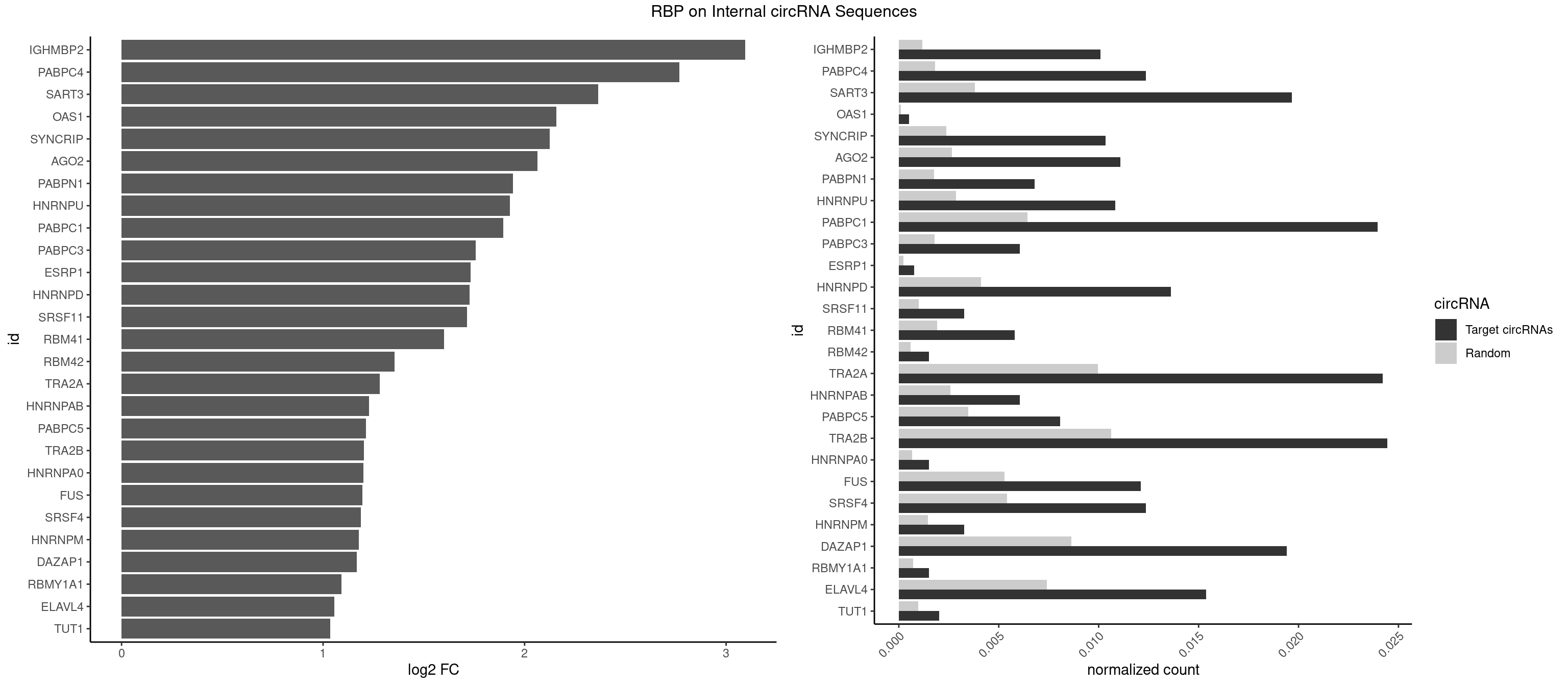
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGHMBP2 | 39 | 813 | 0.0100831863 | 0.0011796520 | 3.095518 | AAAAAA | AAAAAA |
| PABPC4 | 48 | 1251 | 0.0123519032 | 0.0018144033 | 2.767166 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| SART3 | 77 | 2634 | 0.0196622133 | 0.0038186524 | 2.364290 | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| OAS1 | 1 | 77 | 0.0005041593 | 0.0001130379 | 2.157073 | GCAUAA | GCAUAA |
| SYNCRIP | 40 | 1634 | 0.0103352659 | 0.0023694485 | 2.124952 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| AGO2 | 43 | 1830 | 0.0110915049 | 0.0026534924 | 2.063491 | AAAAAA,AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPN1 | 26 | 1222 | 0.0068061507 | 0.0017723764 | 1.941154 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| HNRNPU | 42 | 1965 | 0.0108394253 | 0.0028491350 | 1.927692 | AAAAAA,CCCCCC,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| PABPC1 | 94 | 4443 | 0.0239475674 | 0.0064402624 | 1.894688 | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAAUA,CUAACA,CUAAUA,CUAAUC,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| PABPC3 | 23 | 1234 | 0.0060499118 | 0.0017897669 | 1.757142 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ESRP1 | 2 | 156 | 0.0007562390 | 0.0002275250 | 1.732817 | AGGGAU | AGGGAU |
| HNRNPD | 53 | 2837 | 0.0136123015 | 0.0041128408 | 1.726704 | AAAAAA,AAUUUA,AGAUAU,AUUUAU,UUAGAG,UUAGGA,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| SRSF11 | 12 | 688 | 0.0032770355 | 0.0009985015 | 1.714555 | AAGAAG | AAGAAG |
| RBM41 | 22 | 1318 | 0.0057978321 | 0.0019115000 | 1.600808 | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM42 | 5 | 407 | 0.0015124779 | 0.0005912752 | 1.355012 | AACUAA,AACUAC,ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| TRA2A | 95 | 6871 | 0.0241996471 | 0.0099589296 | 1.280923 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| HNRNPAB | 23 | 1782 | 0.0060499118 | 0.0025839306 | 1.227347 | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPC5 | 31 | 2400 | 0.0080665490 | 0.0034795387 | 1.213056 | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TRA2B | 96 | 7329 | 0.0244517267 | 0.0106226650 | 1.202791 | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| HNRNPA0 | 5 | 453 | 0.0015124779 | 0.0006579386 | 1.200889 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| FUS | 47 | 3648 | 0.0120998235 | 0.0052881452 | 1.194152 | AAAAAA,UGGUGA,UGGUGG,UGGUGU,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| SRSF4 | 48 | 3740 | 0.0123519032 | 0.0054214720 | 1.187977 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPM | 12 | 999 | 0.0032770355 | 0.0014492040 | 1.177131 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| DAZAP1 | 76 | 5964 | 0.0194101336 | 0.0086445016 | 1.166955 | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUG,AGUAAA,AGUAUA,AGUAUG,AGUUAA,AGUUUG,UAGGAA,UAGUAA,UAGUAU,UAGUUU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| RBMY1A1 | 5 | 489 | 0.0015124779 | 0.0007101099 | 1.090800 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| ELAVL4 | 60 | 5105 | 0.0153768591 | 0.0073996354 | 1.055235 | AAAAAA,AUCUAA,AUUUAU,UAUCUA,UAUUUU,UCUAAU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| TUT1 | 7 | 678 | 0.0020166373 | 0.0009840095 | 1.035207 | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
RBP on BSJ (Exon Only)
Plot
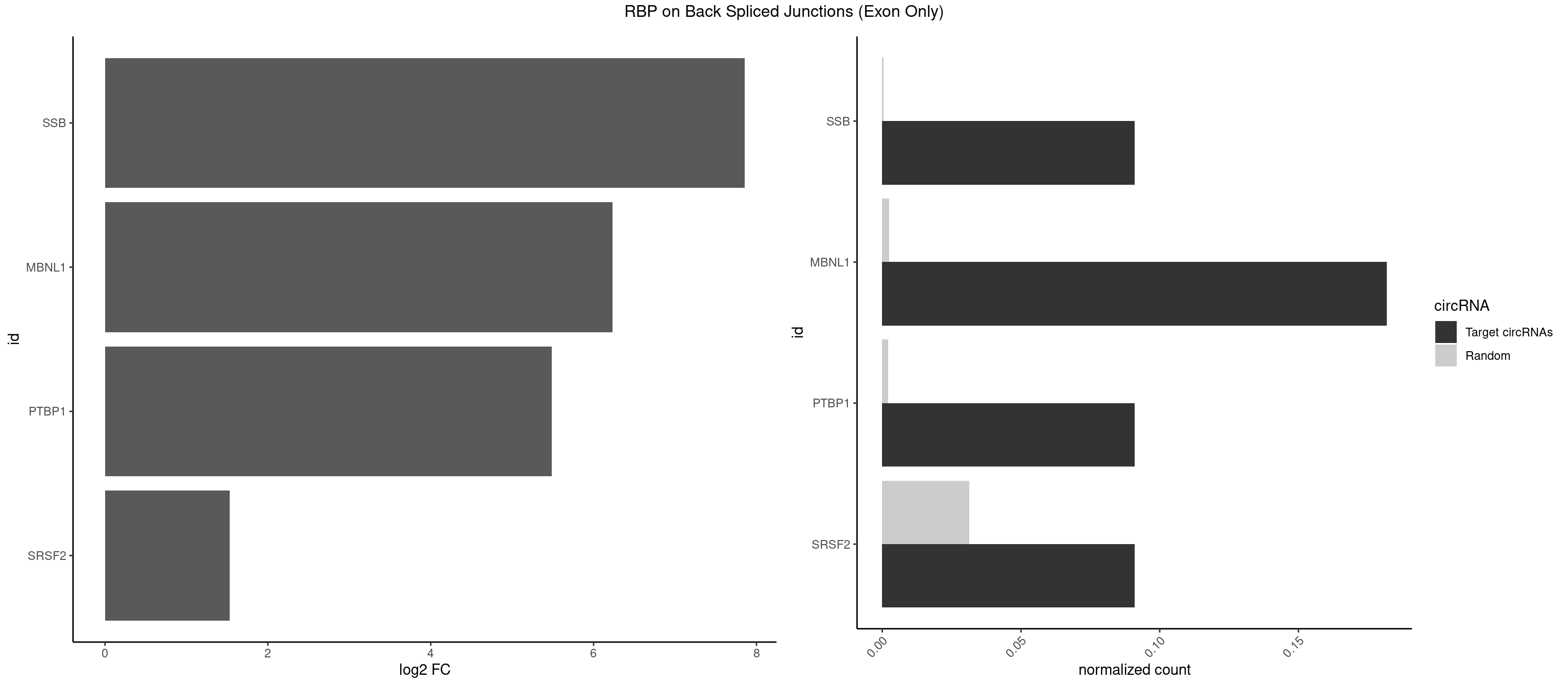
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SSB | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | UGCUGU | CUGUUU,GCUGUU,UGCUGU |
| MBNL1 | 3 | 36 | 0.18181818 | 0.0024233691 | 6.229338 | UGCUGU,UUGCUG,UUUGCU | AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUG,CCUGCU,CGCUGC,CGCUGU,CUGCCG,CUGCCU,CUGCGG,CUGCUC,CUGCUU,CUUGCU,GCUUGC,GCUUGU,GGCUUU,GUGCUG,UGCGGC,UGCUGC,UGCUGU,UGCUUU,UUGCCU,UUGCUC,UUGCUG,UUGCUU,UUGUGC |
| PTBP1 | 1 | 30 | 0.09090909 | 0.0020303904 | 5.484596 | GCUGUG | AGCUGU,CAUCUU,CCAUCU,CCUCUU,CUAUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUUCU,GGCUCC,GUCUUU,UACUUU,UAUUUU,UCCUCU,UCUAUC,UCUCUU,UCUUCU,UCUUUC,UUACUU,UUCUUC,UUCUUG,UUUCUU |
| SRSF2 | 1 | 479 | 0.09090909 | 0.0314383023 | 1.531901 | UGCUGU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
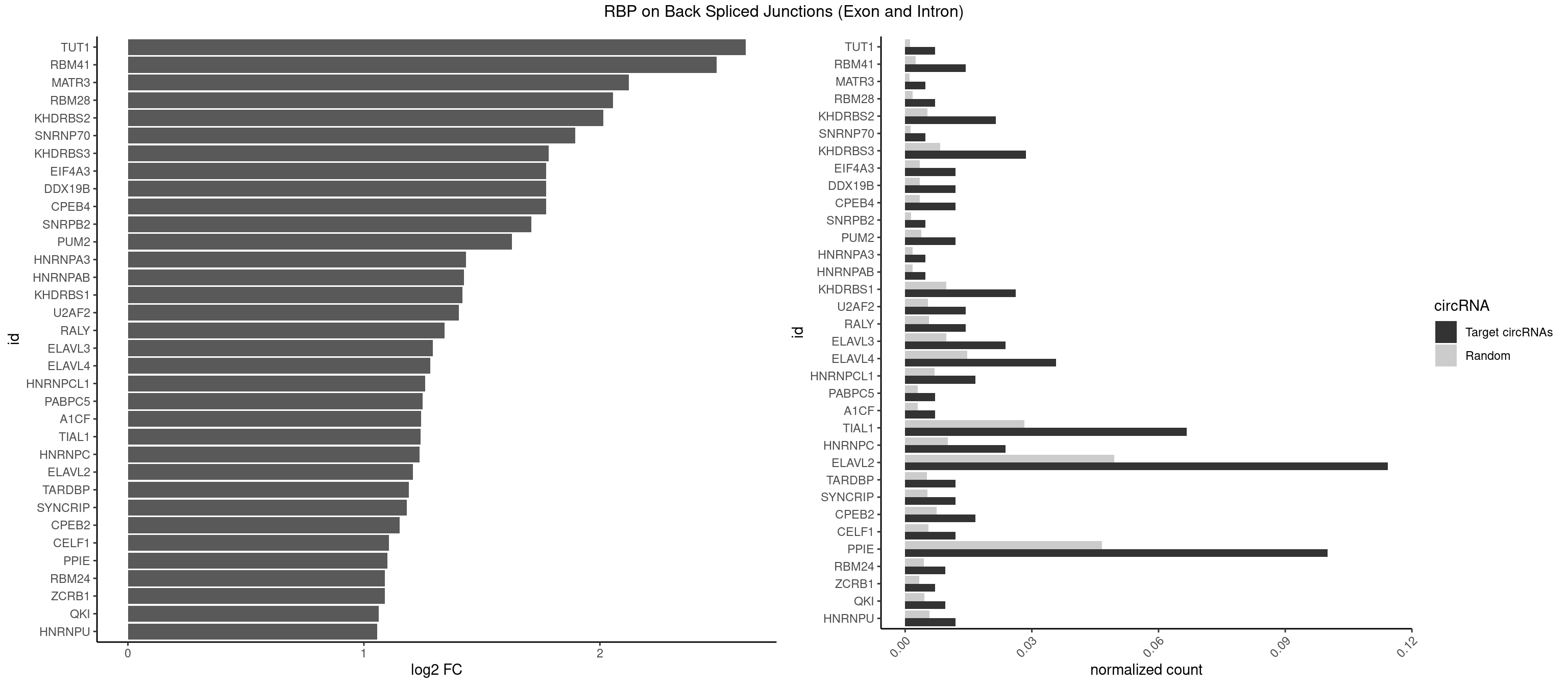
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TUT1 | 2 | 230 | 0.007142857 | 0.001163845 | 2.617602 | AAUACU,CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM41 | 5 | 502 | 0.014285714 | 0.002534260 | 2.494937 | AUACUU,UACAUG,UACUUG,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| MATR3 | 1 | 216 | 0.004761905 | 0.001093309 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM28 | 2 | 340 | 0.007142857 | 0.001718057 | 2.055723 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| KHDRBS2 | 8 | 1051 | 0.021428571 | 0.005300282 | 2.015395 | AAUAAA,AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.001279726 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| KHDRBS3 | 11 | 1646 | 0.028571429 | 0.008298065 | 1.783726 | AAAUAA,AAUAAA,AUAAAA,AUAAAG,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| CPEB4 | 4 | 691 | 0.011904762 | 0.003486497 | 1.771688 | UUUUUU | UUUUUU |
| DDX19B | 4 | 691 | 0.011904762 | 0.003486497 | 1.771688 | UUUUUU | UUUUUU |
| EIF4A3 | 4 | 691 | 0.011904762 | 0.003486497 | 1.771688 | UUUUUU | UUUUUU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.001456066 | 1.709463 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PUM2 | 4 | 764 | 0.011904762 | 0.003854293 | 1.627001 | UACAUA,UGUAGA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.001763402 | 1.433177 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.001773478 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| KHDRBS1 | 10 | 1945 | 0.026190476 | 0.009804514 | 1.417524 | AUAAAA,CUAAAU,UAAAAA,UUAAAU,UUUUAC,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| U2AF2 | 5 | 1071 | 0.014285714 | 0.005401048 | 1.403262 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RALY | 5 | 1117 | 0.014285714 | 0.005632809 | 1.342647 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ELAVL3 | 9 | 1928 | 0.023809524 | 0.009718863 | 1.292679 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ELAVL4 | 14 | 2916 | 0.035714286 | 0.014696695 | 1.281010 | AUUUAU,UAUUUA,UAUUUU,UUGUAU,UUUGUA,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPCL1 | 6 | 1381 | 0.016666667 | 0.006962918 | 1.259202 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.003007860 | 1.247764 | AGAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.003017936 | 1.242939 | AGUAUA,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| TIAL1 | 27 | 5597 | 0.066666667 | 0.028204353 | 1.241048 | AAAUUU,AAUUUU,AUUUUA,AUUUUC,AUUUUU,CAGUUU,CUUUUG,CUUUUU,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUCAGU,UUUUAA,UUUUUA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPC | 9 | 2006 | 0.023809524 | 0.010111850 | 1.235492 | AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL2 | 47 | 9826 | 0.114285714 | 0.049511286 | 1.206816 | AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAU,AUUUUA,AUUUUC,AUUUUU,CUUUUU,GAUUUU,GUUUUU,UAUACU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAUAU,UUCUUU,UUGUAU,UUUAAG,UUUAUA,UUUCUU,UUUGAU,UUUUAA,UUUUUA,UUUUUC,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.005214631 | 1.190902 | GAAUGG,GAAUGU,GUGUGU,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SYNCRIP | 4 | 1041 | 0.011904762 | 0.005249899 | 1.181177 | UUUUUU | AAAAAA,UUUUUU |
| CPEB2 | 6 | 1487 | 0.016666667 | 0.007496977 | 1.152585 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| CELF1 | 4 | 1097 | 0.011904762 | 0.005532044 | 1.105654 | GUGUGU,GUUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| PPIE | 41 | 9262 | 0.100000000 | 0.046669690 | 1.099442 | AAAAAU,AAAUAA,AAAUUU,AAUAAA,AAUUUU,AUAAAA,AUAAAU,AUAUUU,AUUAUU,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUU,UAUAAA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAU,UUAUAU,UUUAUA,UUUUAA,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| RBM24 | 3 | 888 | 0.009523810 | 0.004479041 | 1.088349 | AGUGUG,GAGUGU,GUGUGU | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.003360540 | 1.087808 | ACUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| QKI | 3 | 904 | 0.009523810 | 0.004559653 | 1.062615 | ACUAAU,AUUAAC,CACUAA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPU | 4 | 1136 | 0.011904762 | 0.005728537 | 1.055300 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.