circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000181163:+:5:171391705:171400925
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000181163:+:5:171391705:171400925 | ENSG00000181163 | ENST00000676625 | + | 5 | 171391706 | 171400925 | 411 | GUUUCCCUUGGGGGCUUUGAAAUAACACCACCAGUGGUCUUAAGGUUGAAGUGUGGUUCAGGGCCAGUGCAUAUUAGUGGACAGCACUUAGUAGCUGUGGAGGAAGAUGCAGAGUCAGAAGAUGAAGAGGAGGAGGAUGUGAAACUCUUAAGUAUAUCUGGAAAGCGGUCUGCCCCUGGAGGUGGUAGCAAGGUUCCACAGAAAAAAGUAAAACUUGCUGCUGAUGAAGAUGAUGACGAUGAUGAUGAAGAGGAUGAUGAUGAAGAUGAUGAUGAUGAUGAUUUUGAUGAUGAGGAAGCUGAAGAAAAAGCGCCAGUGAAGAAAUCUAUACGAGAUACUCCAGCCAAAAAUGCACAAAAGUCAAAUCAGAAUGGAAAAGACUCAAAACCAUCAUCAACACCAAGAUCAAAAGUUUCCCUUGGGGGCUUUGAAAUAACACCACCAGUGGUCUUAAGGUUGAA | circ |
| ENSG00000181163:+:5:171391705:171400925 | ENSG00000181163 | ENST00000676625 | + | 5 | 171391706 | 171400925 | 22 | CAAGAUCAAAAGUUUCCCUUGG | bsj |
| ENSG00000181163:+:5:171391705:171400925 | ENSG00000181163 | ENST00000676625 | + | 5 | 171391506 | 171391715 | 210 | AAGGAGAUAGAAAGUGGUUCUUUAUCUUCUGUCACUGGAGUUCGAUGGUCAACUCUUGAACAUGGGGGCUUCUGCUGCUACUUUUAUCAGAGGUGGAAAAACAGGUUCACUGGUUUGUUGAUUUGGCUUAUGUGUUUGCCUGUAAUGUUUAUUGUUCAUUUUCUUCACAUGUUUAGUGAUGAAAAAUUUCUCCCUUCUAGGUUUCCCUUG | ie_up |
| ENSG00000181163:+:5:171391705:171400925 | ENSG00000181163 | ENST00000676625 | + | 5 | 171400916 | 171401125 | 210 | AAGAUCAAAAGUAAGUGGCUACAUUUACACGUGGGUCUCAUUGAUCUAGUUGGGGAAAAAGAUUCUACUGUGGAAGAAUCUAGUGUGUCUGAAAUUUGAUAGGCCUUUAUAGAACCCCUGUAAUUGCUGUUUAAAAGUUAAAAUCAGCUUGCUGCAGCCAGGCUCAGUGGCUCACUCCUGUAAUCCCAGCACUUUGGGAGGGAGGCCAAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
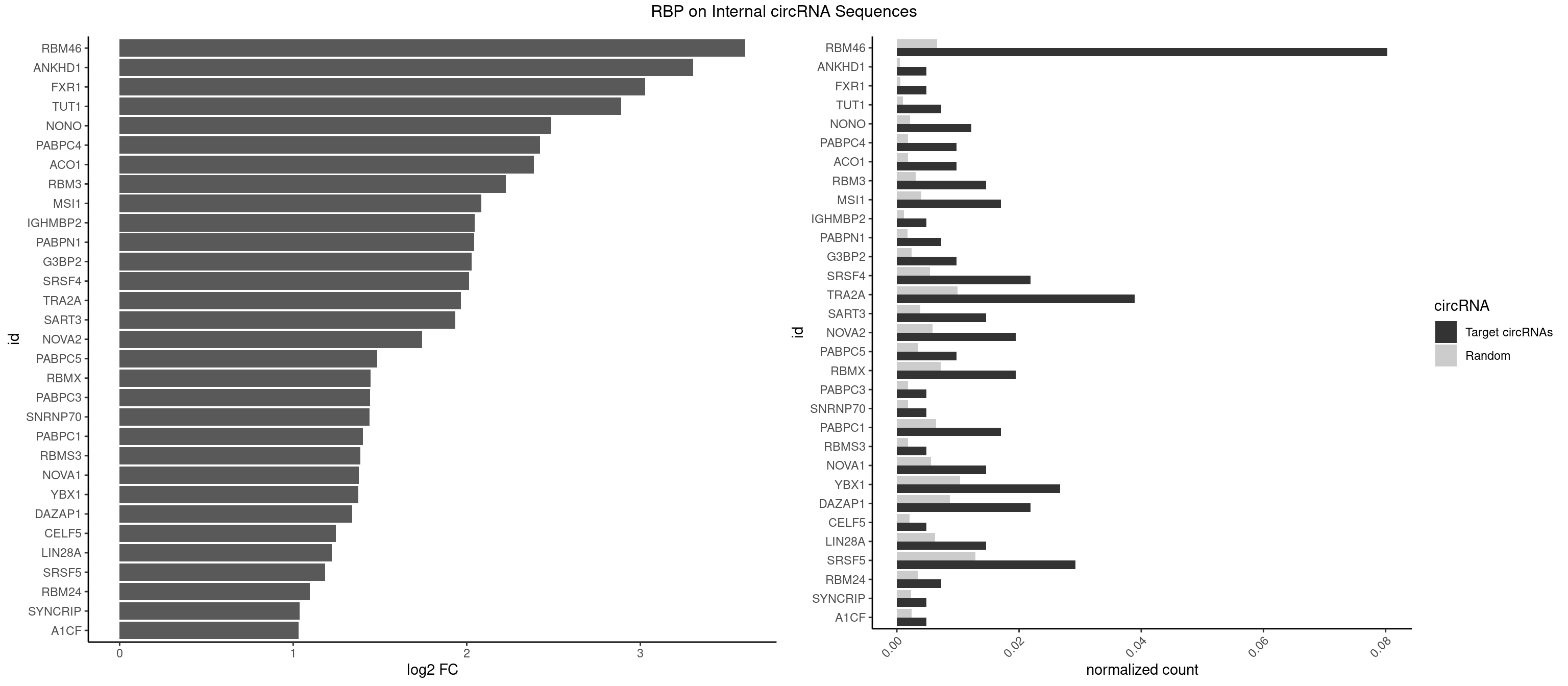
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM46 | 32 | 4554 | 0.08029197 | 0.0066011240 | 3.604472 | AUCAAA,AUGAAG,AUGAUG,AUGAUU,GAUCAA,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ANKHD1 | 1 | 339 | 0.00486618 | 0.0004927293 | 3.303922 | GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| FXR1 | 1 | 411 | 0.00486618 | 0.0005970720 | 3.026813 | AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| TUT1 | 2 | 678 | 0.00729927 | 0.0009840095 | 2.891008 | AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| NONO | 4 | 1498 | 0.01216545 | 0.0021723567 | 2.485457 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PABPC4 | 3 | 1251 | 0.00973236 | 0.0018144033 | 2.423294 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| ACO1 | 3 | 1283 | 0.00973236 | 0.0018607779 | 2.386884 | CAGUGA,CAGUGC,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM3 | 5 | 2152 | 0.01459854 | 0.0031201361 | 2.226143 | AAAACU,AAGACU,AUACGA,GAAACU,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| MSI1 | 6 | 2770 | 0.01703163 | 0.0040157442 | 2.084477 | AGGAAG,AGGAGG,AGGUGG,UAGUAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| IGHMBP2 | 1 | 813 | 0.00486618 | 0.0011796520 | 2.044428 | AAAAAA | AAAAAA |
| PABPN1 | 2 | 1222 | 0.00729927 | 0.0017723764 | 2.042067 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| G3BP2 | 3 | 1644 | 0.00973236 | 0.0023839405 | 2.029441 | AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SRSF4 | 8 | 3740 | 0.02189781 | 0.0054214720 | 2.014030 | AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| TRA2A | 15 | 6871 | 0.03892944 | 0.0099589296 | 1.966799 | AAGAAA,AAGAGG,AGAAGA,AGAGGA,AGGAAG,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SART3 | 5 | 2634 | 0.01459854 | 0.0038186524 | 1.934689 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| NOVA2 | 7 | 4013 | 0.01946472 | 0.0058171047 | 1.742488 | AACACC,ACCACC,AGAUCA,AUCAAC,AUCAUC,GAGUCA | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| PABPC5 | 3 | 2400 | 0.00973236 | 0.0034795387 | 1.483894 | AGAAAA,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBMX | 7 | 4925 | 0.01946472 | 0.0071387787 | 1.447112 | AAGUAA,AAGUGU,AGGAAG,AUCAAA,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| PABPC3 | 1 | 1234 | 0.00486618 | 0.0017897669 | 1.443018 | AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SNRNP70 | 1 | 1237 | 0.00486618 | 0.0017941145 | 1.439518 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPC1 | 6 | 4443 | 0.01703163 | 0.0064402624 | 1.403025 | AAAAAA,AGAAAA,CAAAUC,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| RBMS3 | 1 | 1283 | 0.00486618 | 0.0018607779 | 1.386884 | UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| NOVA1 | 5 | 3872 | 0.01459854 | 0.0056127669 | 1.379040 | AACACC,ACCACC,AUCAAC,AUCAUC | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| YBX1 | 10 | 7119 | 0.02676399 | 0.0103183321 | 1.375083 | ACACCA,ACCACC,AUCAUC,CACCAC,CAUCAU,CCACCA,GGUCUG,GUCUGC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| DAZAP1 | 8 | 5964 | 0.02189781 | 0.0086445016 | 1.340932 | AAAAAA,AGGAAG,AGGAUG,AGGUUG,AGUAAA,AGUAUA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| CELF5 | 1 | 1415 | 0.00486618 | 0.0020520728 | 1.245708 | GUGUGG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| LIN28A | 5 | 4315 | 0.01459854 | 0.0062547643 | 1.222797 | GGAGGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SRSF5 | 11 | 8869 | 0.02919708 | 0.0128544391 | 1.183557 | AGAAGA,AGGAAG,CACAGA,GAAGAA,GAGGAA,GGAAGA,UGCAGA,UGCAUA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| RBM24 | 2 | 2357 | 0.00729927 | 0.0034172229 | 1.094928 | AGUGUG,GUGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| SYNCRIP | 1 | 1634 | 0.00486618 | 0.0023694485 | 1.038238 | AAAAAA | AAAAAA,UUUUUU |
| A1CF | 1 | 1642 | 0.00486618 | 0.0023810421 | 1.031197 | AGUAUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
RBP on BSJ (Exon Only)
Plot
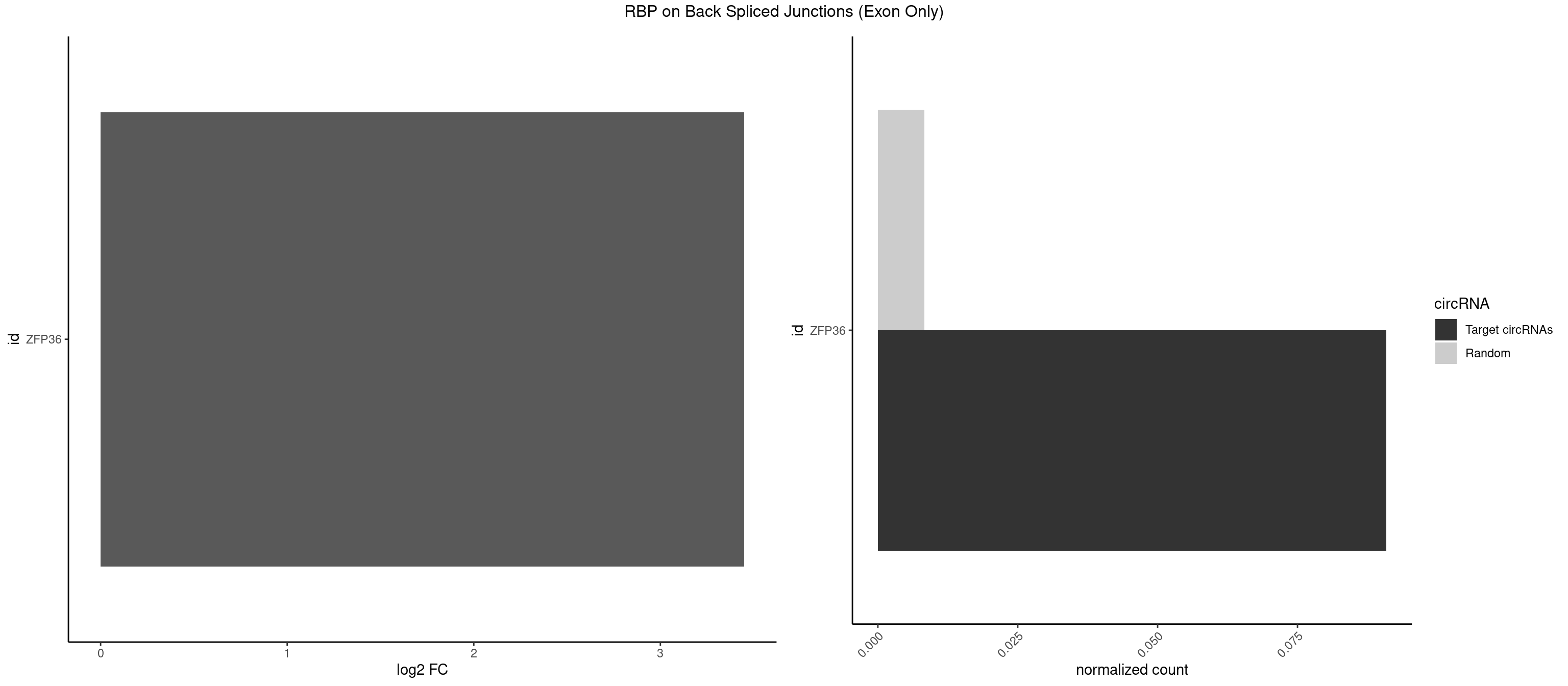
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZFP36 | 1 | 126 | 0.09090909 | 0.008318051 | 3.450107 | AAAAGU | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
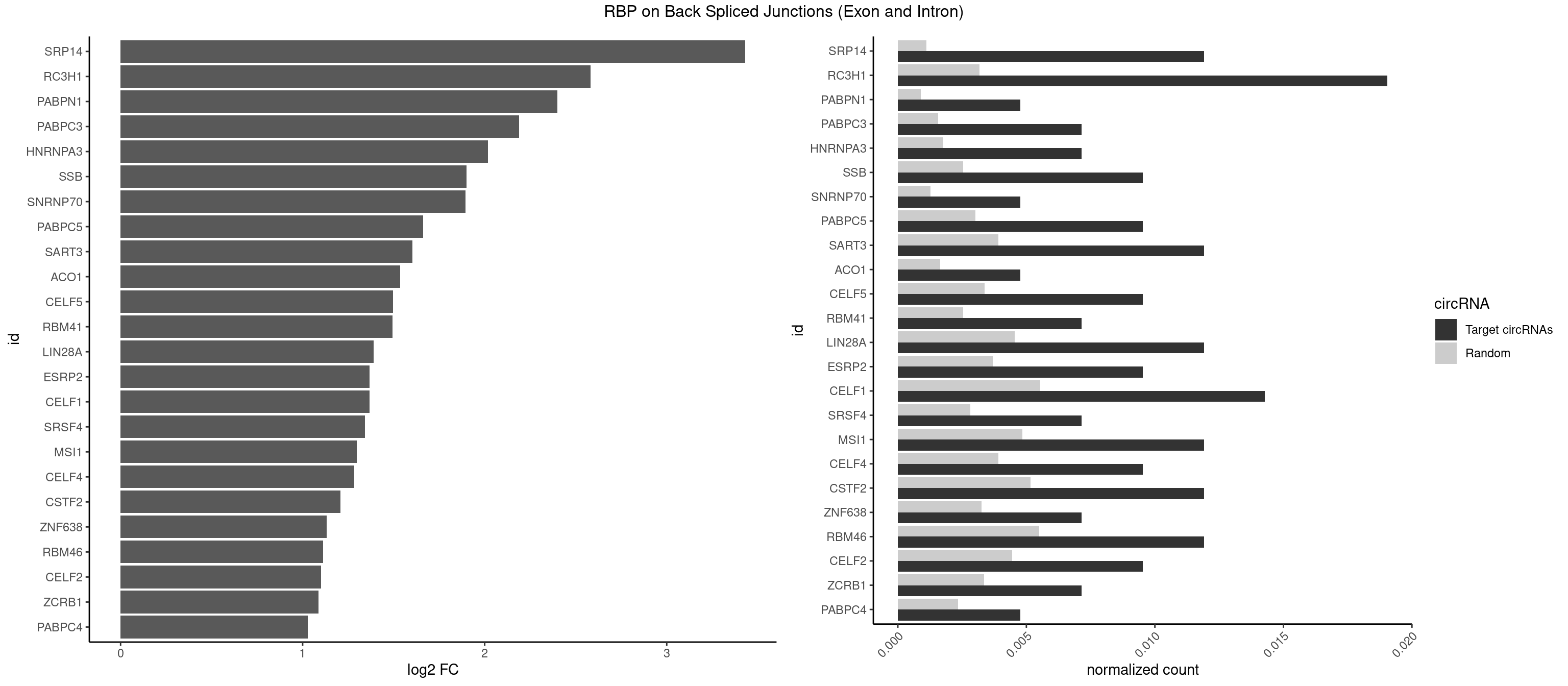
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRP14 | 4 | 218 | 0.011904762 | 0.0011033857 | 3.431530 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RC3H1 | 7 | 631 | 0.019047619 | 0.0031841999 | 2.580608 | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AAGGAG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAG,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SART3 | 4 | 777 | 0.011904762 | 0.0039197904 | 1.602690 | AAAAAC,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGU,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACAUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | AGGAGA,GGAGAU,GGAGGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| CELF1 | 5 | 1097 | 0.014285714 | 0.0055320435 | 1.368689 | GUGUGU,GUUUGU,UGUCUG,UGUGUU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGUGG,AGUAAG,AGUUGG,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| CELF4 | 3 | 776 | 0.009523810 | 0.0039147521 | 1.282618 | GUGUGU,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUGU,GUGUUU,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU,GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AUCAAA,AUGAAA,GAUCAA,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | AUGUGU,GUGUGU,UAUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GGCUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.