circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000130816:-:19:10177291:10182077
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000130816:-:19:10177291:10182077 | ENSG00000130816 | ENST00000676610 | - | 19 | 10177292 | 10182077 | 441 | GCUCAAAGAUUUGGAAAGAGACAGCUUAACAGAAAAGGAAUGUGUGAAGGAGAAAUUGAAUCUCUUGCACGAAUUUCUGCAAACAGAAAUAAAGAAUCAGUUAUGUGACUUGGAAACCAAAUUACGUAAAGAAGAAUUAUCCGAGGAGGGCUACCUGGCUAAAGUCAAAUCCCUUUUAAAUAAAGAUUUGUCCUUGGAGAACGGUGCUCAUGCUUACAACCGGGAAGUGAAUGGACGUCUAGAAAACGGGAACCAAGCAAGAAGUGAAGCCCGUAGAGUGGGAAUGGCAGAUGCCAACAGCCCCCCCAAACCCCUUUCCAAACCUCGCACGCCCAGGAGGAGCAAGUCCGAUGGAGAGGCUAAGCCUGAACCUUCACCUAGCCCCAGGAUUACAAGGAAAAGCACCAGGCAAACCACCAUCACAUCUCAUUUUGCAAAGGGGCUCAAAGAUUUGGAAAGAGACAGCUUAACAGAAAAGGAAUGUGUGAAGG | circ |
| ENSG00000130816:-:19:10177291:10182077 | ENSG00000130816 | ENST00000676610 | - | 19 | 10177292 | 10182077 | 22 | UUUGCAAAGGGGCUCAAAGAUU | bsj |
| ENSG00000130816:-:19:10177291:10182077 | ENSG00000130816 | ENST00000676610 | - | 19 | 10182068 | 10182277 | 210 | UCAGAUGACUUCUUAGCAUUAGGCAUUCCAGUAGGACACUCUAGACUCUUGCGGGGAGACAAAAGCCAGCUUAGUUUUUUCUAACACUCAUAUGUUAAACUUGUUUGUGUCCAAAACUUCUUUAGAACUGUGAUAUUCUUACAGGCAAAUGAAGUUGCUUAACAAGUGUUUGUAUUUUCUCCCCUAUUUCUUCCUCCCAGGCUCAAAGAU | ie_up |
| ENSG00000130816:-:19:10177291:10182077 | ENSG00000130816 | ENST00000676610 | - | 19 | 10177092 | 10177301 | 210 | UUGCAAAGGGGUCAGUAUACGAUAAAUUGGCGGCUGCCUUUUUUAGGGGCCGGCUGUUUUGGGAUGGAAUUGGUAGGGCGUCACGUGGCAAUUCUGUCUUCCGUGUUGUAUAGGUUUUCAUCUUGCGGUUCUGUUCUCUAGACAUUUAUGAAUAGCAUUUCUGAAAACGUUACCUGUUUUUACUCAUAUAGGAAAGUUCAAAUGUACACA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
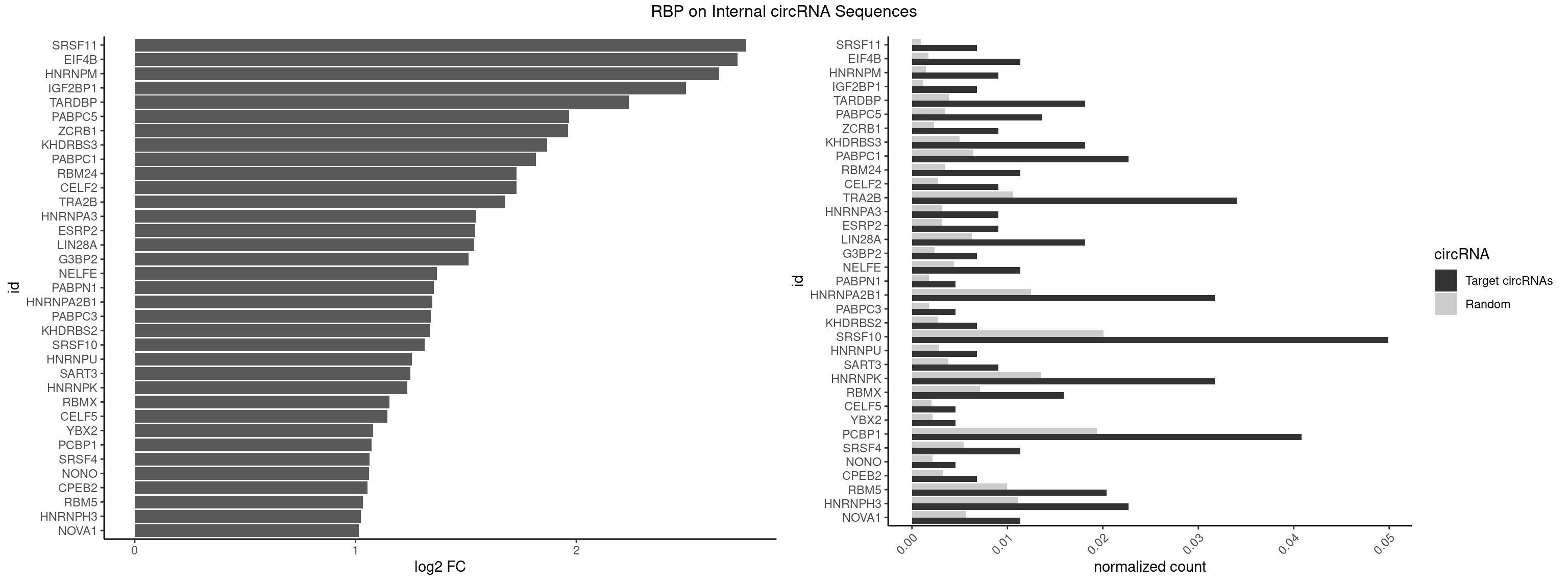
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF11 | 2 | 688 | 0.006802721 | 0.0009985015 | 2.768275 | AAGAAG | AAGAAG |
| EIF4B | 4 | 1179 | 0.011337868 | 0.0017100607 | 2.729030 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPM | 3 | 999 | 0.009070295 | 0.0014492040 | 2.645889 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| IGF2BP1 | 2 | 831 | 0.006802721 | 0.0012057377 | 2.496196 | AAGCAC,AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| TARDBP | 7 | 2654 | 0.018140590 | 0.0038476365 | 2.237177 | GAAUGG,GAAUGU,GUGAAU,UGAAUG,UGUGUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| PABPC5 | 5 | 2400 | 0.013605442 | 0.0034795387 | 1.967216 | AGAAAA,AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZCRB1 | 3 | 1605 | 0.009070295 | 0.0023274215 | 1.962417 | GAAUUA,GCUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| KHDRBS3 | 7 | 3429 | 0.018140590 | 0.0049707696 | 1.867680 | AAAUAA,AAUAAA,AUAAAG,UAAAUA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| PABPC1 | 9 | 4443 | 0.022675737 | 0.0064402624 | 1.815958 | ACGAAU,AGAAAA,CAAACA,CAAACC,CAAAUC,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| RBM24 | 4 | 2357 | 0.011337868 | 0.0034172229 | 1.730253 | AGAGUG,GAGUGG,GUGUGA,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF2 | 3 | 1886 | 0.009070295 | 0.0027346479 | 1.729794 | AUGUGU,UAUGUG,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| TRA2B | 14 | 7329 | 0.034013605 | 0.0106226650 | 1.678966 | AAAGAA,AAGAAG,AAGAAU,AAGGAA,AGAAGA,AGGAAA,GAAAGA,GAAGAA,GAAGGA,GAAUUA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| HNRNPA3 | 3 | 2140 | 0.009070295 | 0.0031027457 | 1.547604 | AAGGAG,AGGAGC,CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 3 | 2150 | 0.009070295 | 0.0031172377 | 1.540881 | GGGAAG,GGGAAU,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| LIN28A | 7 | 4315 | 0.018140590 | 0.0062547643 | 1.536194 | AGGAGA,GGAGAA,GGAGGA,GGAGGG,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| G3BP2 | 2 | 1644 | 0.006802721 | 0.0023839405 | 1.512764 | AGGAUU,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| NELFE | 4 | 3028 | 0.011337868 | 0.0043896388 | 1.368975 | CUGGCU,GGCUAA,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| PABPN1 | 1 | 1222 | 0.004535147 | 0.0017723764 | 1.355464 | AGAAGA | AAAAGA,AGAAGA |
| HNRNPA2B1 | 13 | 8607 | 0.031746032 | 0.0124747476 | 1.347566 | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AGGAGC,AGGGGC,CAAGAA,CAAGGA,GAAGCC,GAAGGA,GGAACC | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| PABPC3 | 1 | 1234 | 0.004535147 | 0.0017897669 | 1.341378 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| KHDRBS2 | 2 | 1858 | 0.006802721 | 0.0026940701 | 1.336325 | AAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| SRSF10 | 21 | 13860 | 0.049886621 | 0.0200874160 | 1.312361 | AAAGAA,AAAGAG,AAAGGA,AAAGGG,AAGAAG,AAGAGA,AAGGAA,AAGGAG,AAGGGG,AGAGAC,CAAAGA,GAAAGA,GAGAAA,GAGAAC,GAGACA,GAGAGG,GAGGAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| HNRNPU | 2 | 1965 | 0.006802721 | 0.0028491350 | 1.255588 | CCCCCC | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| SART3 | 3 | 2634 | 0.009070295 | 0.0038186524 | 1.248086 | AGAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| HNRNPK | 13 | 9304 | 0.031746032 | 0.0134848428 | 1.235238 | CAAACC,CCAAAC,CCCCAA,CCCCCA,CCCCCC,CCCCUU,GCCCAG,GCCCCC,UCCCUU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| RBMX | 6 | 4925 | 0.015873016 | 0.0071387787 | 1.152827 | AAGAAG,AAGGAA,ACCAAA,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| CELF5 | 1 | 1415 | 0.004535147 | 0.0020520728 | 1.144068 | UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| YBX2 | 1 | 1480 | 0.004535147 | 0.0021462711 | 1.079317 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PCBP1 | 17 | 13384 | 0.040816327 | 0.0193975949 | 1.073269 | AAACCA,AAAUUA,AACCAA,AAUUAC,ACCAAA,CAAACC,CCAAAC,CCCCAA,CCCCCC,CCUUUC,CUUUCC | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| SRSF4 | 4 | 3740 | 0.011337868 | 0.0054214720 | 1.064393 | AAGAAG,AGAAGA,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| NONO | 1 | 1498 | 0.004535147 | 0.0021723567 | 1.061888 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| CPEB2 | 2 | 2261 | 0.006802721 | 0.0032780993 | 1.053252 | CAUUUU,CCUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RBM5 | 8 | 6879 | 0.020408163 | 0.0099705232 | 1.033405 | AAGGAA,AAGGAG,AAGGGG,CAAGGA,CCCCCC,GAAGGA | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| HNRNPH3 | 9 | 7688 | 0.022675737 | 0.0111429292 | 1.025021 | AAUGUG,GAAUGU,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGGCU | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| NOVA1 | 4 | 3872 | 0.011337868 | 0.0056127669 | 1.014365 | ACCACC,AGCACC,CCCCCC | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
RBP on BSJ (Exon Only)
Plot
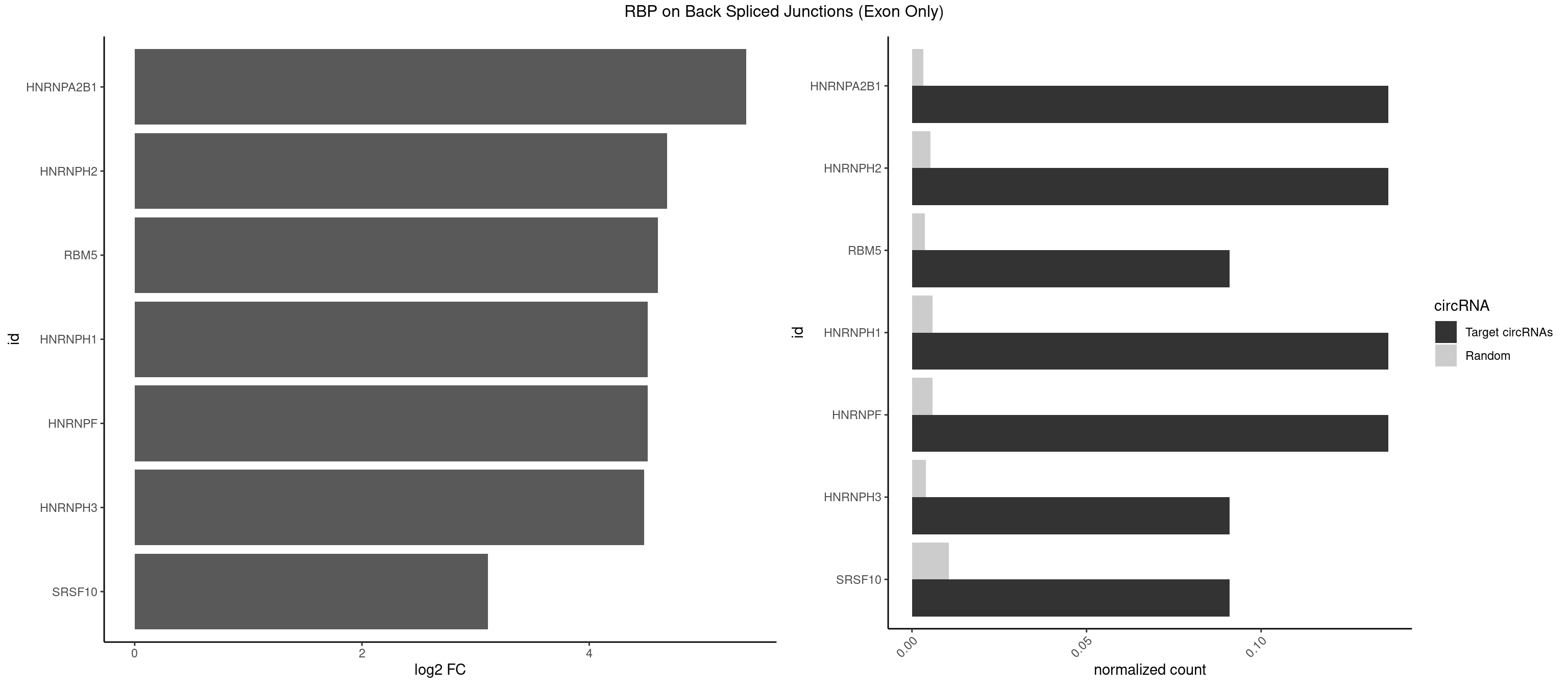
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA2B1 | 2 | 49 | 0.13636364 | 0.003274823 | 5.379898 | AAGGGG,AGGGGC | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| HNRNPH2 | 2 | 80 | 0.13636364 | 0.005305214 | 4.683904 | AAGGGG,GGGGCU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| RBM5 | 1 | 56 | 0.09090909 | 0.003733298 | 4.605902 | AAGGGG | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
| HNRNPF | 2 | 90 | 0.13636364 | 0.005960178 | 4.515960 | AAGGGG,GGGGCU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 2 | 90 | 0.13636364 | 0.005960178 | 4.515960 | AAGGGG,GGGGCU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPH3 | 1 | 61 | 0.09090909 | 0.004060781 | 4.484596 | GGGGCU | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| SRSF10 | 1 | 160 | 0.09090909 | 0.010544931 | 3.107875 | AAGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon and Intron)
Plot
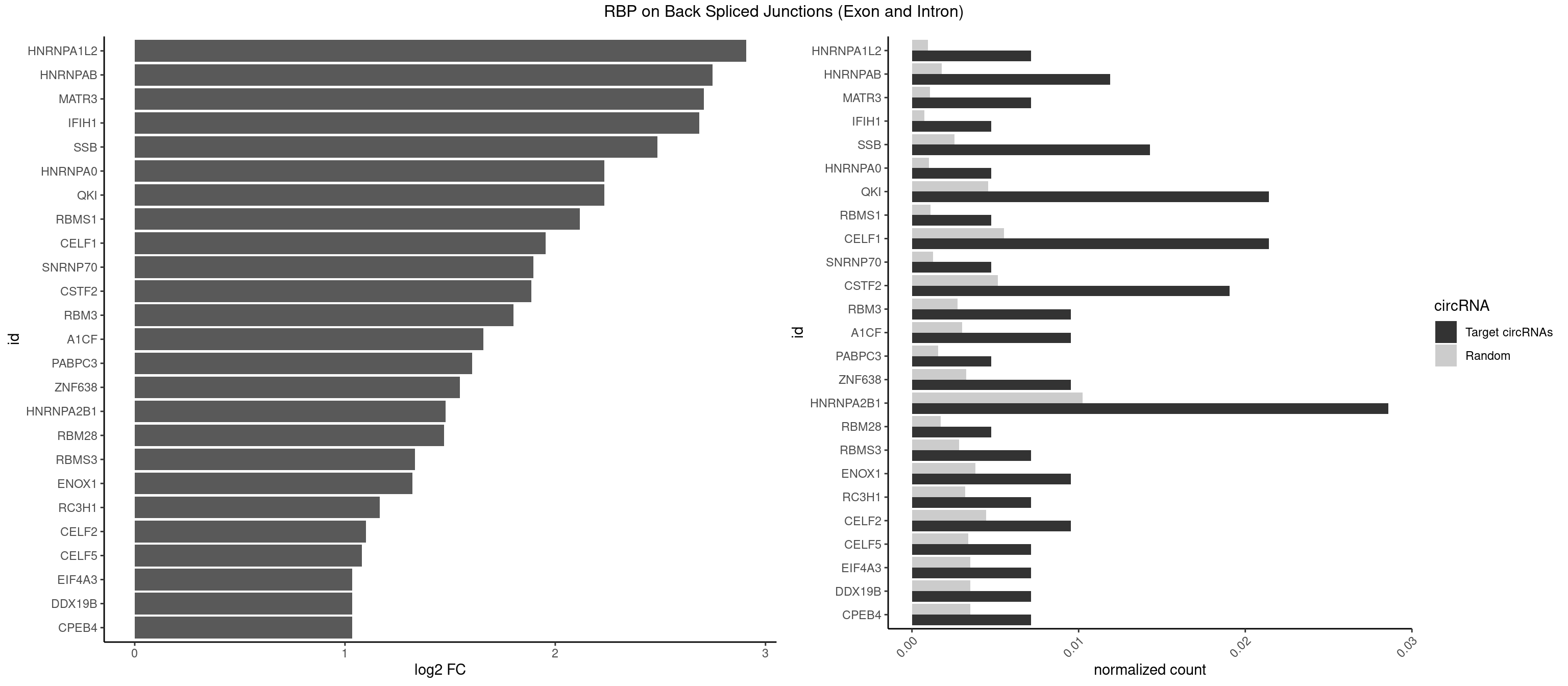
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | GUAGGG,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPAB | 4 | 351 | 0.011904762 | 0.0017734784 | 2.746885 | AGACAA,AUAGCA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| SSB | 5 | 505 | 0.014285714 | 0.0025493753 | 2.486358 | CUGUUU,GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| QKI | 8 | 904 | 0.021428571 | 0.0045596534 | 2.232540 | ACUCAU,CUAACA,CUCAUA,UACUCA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| CELF1 | 8 | 1097 | 0.021428571 | 0.0055320435 | 1.953651 | CUGUCU,GUGUUG,GUUUGU,UGUUGU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CSTF2 | 7 | 1022 | 0.019047619 | 0.0051541717 | 1.885798 | GUGUUG,GUGUUU,GUUUUG,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACG,AAAACU,AUACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | AGUAUA,CAGUAU,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUCU,UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPA2B1 | 11 | 2033 | 0.028571429 | 0.0102478839 | 1.479247 | AAGGGG,AGGGGC,AGUAGG,AUAGCA,CUAGAC,GGGGCC,GUAGGG,UAGACA,UAGACU,UUAGGG | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | AUAUAG,CAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AGGACA,UAGACA,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | UAUGUU,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUG,GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.