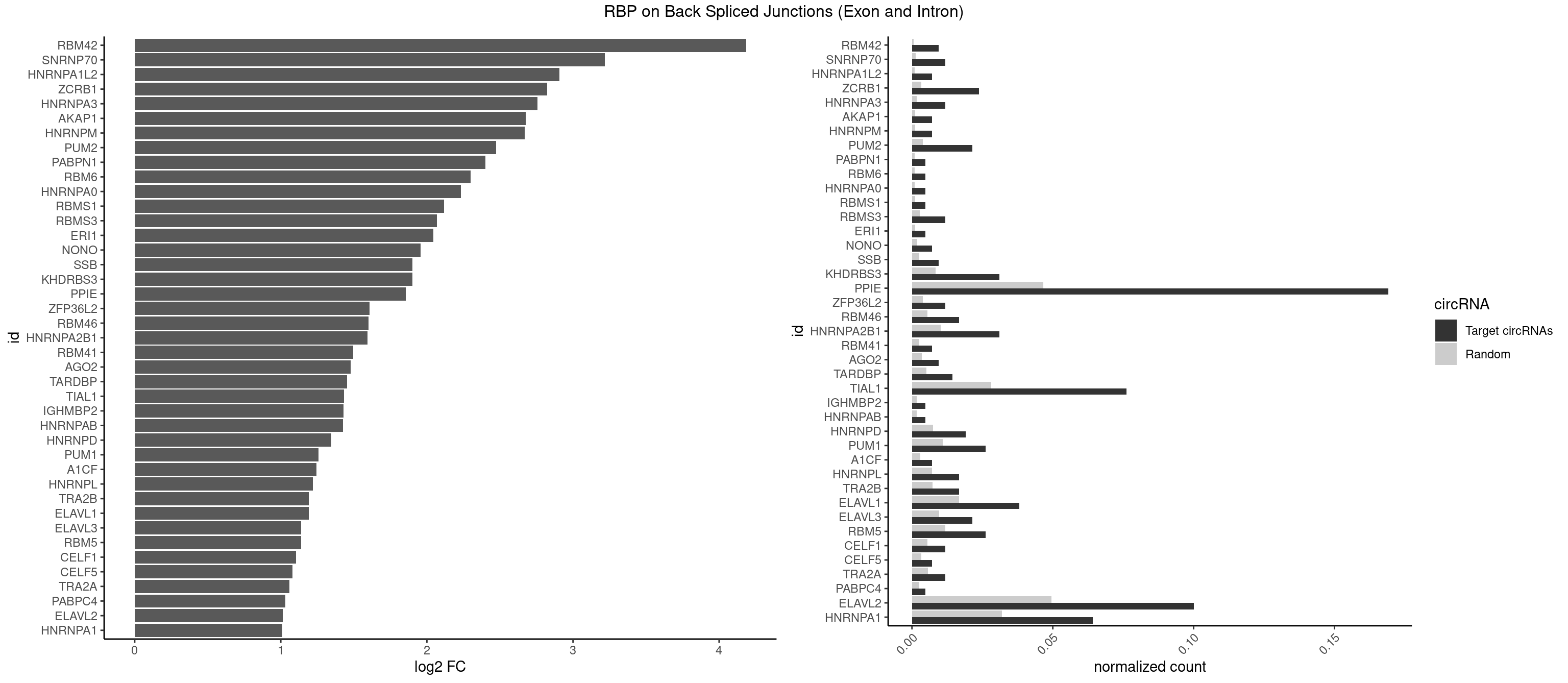
| RBM42 |
3 |
103 |
0.009523810 |
0.0005239823 |
4.183949 |
AACUAA,ACUAAG |
AACUAA,AACUAC,ACUAAG,ACUACG |
| SNRNP70 |
4 |
253 |
0.011904762 |
0.0012797259 |
3.217632 |
AAUCAA,AUCAAG,GUUCAA,UUCAAG |
AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPA1L2 |
2 |
188 |
0.007142857 |
0.0009522370 |
2.907109 |
AUAGGG,UAGGGU |
AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| ZCRB1 |
9 |
666 |
0.023809524 |
0.0033605401 |
2.824774 |
AAUUAA,ACUUAA,AUUUAA,GACUUA,GAUUAA,GAUUUA,GCUUAA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPA3 |
4 |
349 |
0.011904762 |
0.0017634019 |
2.755106 |
AAGGAG,CAAGGA,CCAAGG,GCCAAG |
AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| AKAP1 |
2 |
221 |
0.007142857 |
0.0011185006 |
2.674935 |
AUAUAU,UAUAUA |
AUAUAU,UAUAUA |
| HNRNPM |
2 |
222 |
0.007142857 |
0.0011235389 |
2.668451 |
AAGGAA,GAAGGA |
AAGGAA,GAAGGA,GGGGGG |
| PUM2 |
8 |
764 |
0.021428571 |
0.0038542926 |
2.474998 |
GUAAAU,UAAAUA,UACAUA,UAGAUA,UAUAUA,UGUAAA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPN1 |
1 |
178 |
0.004761905 |
0.0009018541 |
2.400573 |
AAAAGA |
AAAAGA,AGAAGA |
| RBM6 |
1 |
191 |
0.004761905 |
0.0009673519 |
2.299426 |
AAUCCA |
AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPA0 |
1 |
200 |
0.004761905 |
0.0010126965 |
2.233337 |
AGAUAU |
AAUUUA,AGAUAU,AGUAGG |
| RBMS1 |
1 |
217 |
0.004761905 |
0.0010983474 |
2.116204 |
UAUAUA |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBMS3 |
4 |
562 |
0.011904762 |
0.0028365578 |
2.069326 |
AAUAUA,AUAUAU,UAUAUA |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ERI1 |
1 |
228 |
0.004761905 |
0.0011537686 |
2.045185 |
UUCAGA |
UUCAGA,UUUCAG |
| NONO |
2 |
364 |
0.007142857 |
0.0018389762 |
1.957598 |
AGAGGA,GAGGAA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SSB |
3 |
505 |
0.009523810 |
0.0025493753 |
1.901395 |
GCUGUU,UGCUGU,UGUUUU |
CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| KHDRBS3 |
12 |
1646 |
0.030952381 |
0.0082980653 |
1.899203 |
AAAUAA,AAUAAA,AUAAAC,AUAAAG,AUUAAA,UAAAUA,UUAAAC |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
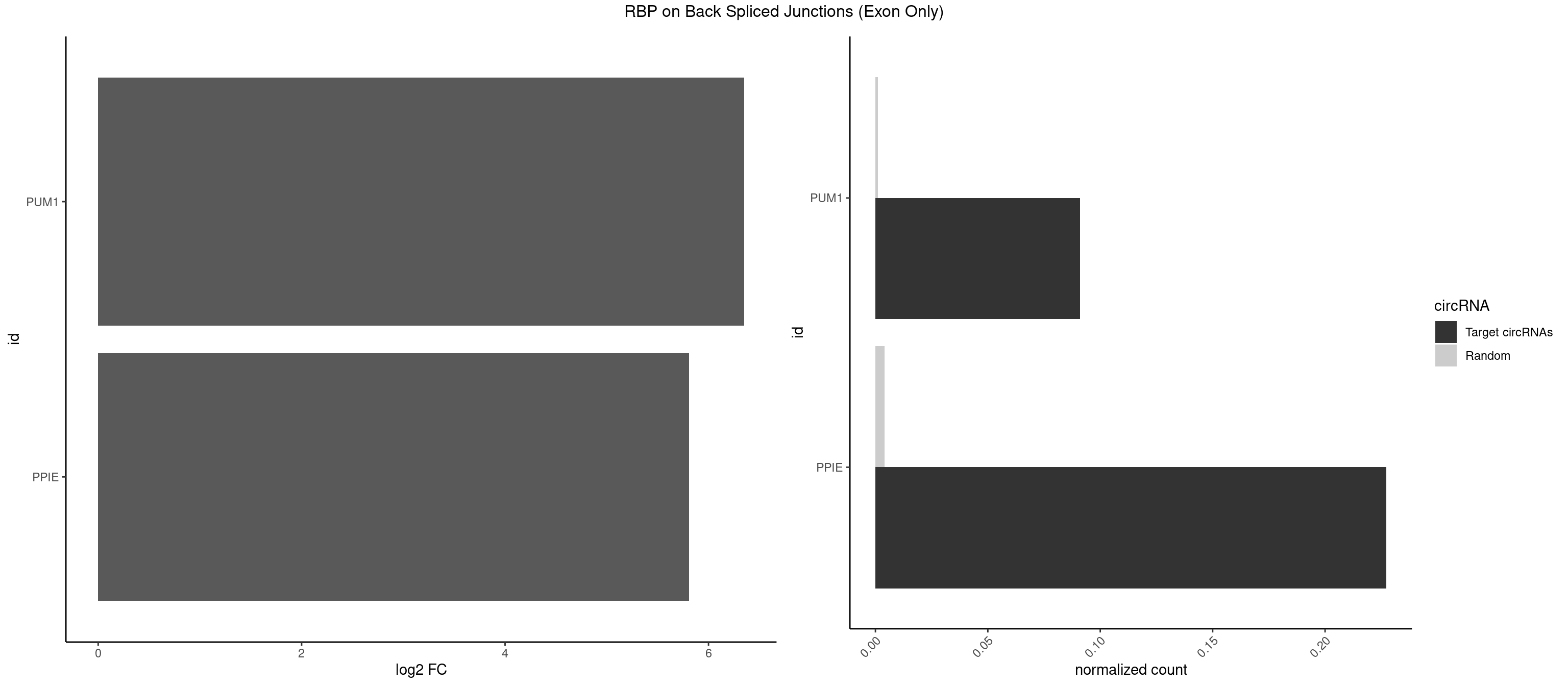
| PPIE |
70 |
9262 |
0.169047619 |
0.0466696896 |
1.856872 |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUUAA,AAUUUU,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUU,AUUAAA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAU,UAAAUA,UAAAUU,UAAUUU,UAUAAA,UAUAUA,UAUUAA,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUU,UUAUAA,UUAUUA,UUAUUU,UUUAAA,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| ZFP36L2 |
4 |
774 |
0.011904762 |
0.0039046755 |
1.608264 |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM46 |
6 |
1091 |
0.016666667 |
0.0055018138 |
1.598986 |
AAUCAA,AAUGAA,AUCAAG,AUCAUA,AUCAUU,AUGAAA |
AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPA2B1 |
12 |
2033 |
0.030952381 |
0.0102478839 |
1.594724 |
AAGGAA,AAGGAG,AGAUAU,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CCAAGG,GAAGGA,GCCAAG,UAGGGU,UUUAUA |
AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| RBM41 |
2 |
502 |
0.007142857 |
0.0025342604 |
1.494937 |
AUACAU,UUACUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| AGO2 |
3 |
677 |
0.009523810 |
0.0034159613 |
1.479247 |
AAAAAA,AGUGCU,GUGCUU |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| TARDBP |
5 |
1034 |
0.014285714 |
0.0052146312 |
1.453936 |
GAAUGA,GUGAAU,UGAAUG,UUUUGC |
GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| TIAL1 |
31 |
5597 |
0.076190476 |
0.0282043531 |
1.433693 |
AAAUUU,AAUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,GUUUUU,UAAAUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| IGHMBP2 |
1 |
350 |
0.004761905 |
0.0017684401 |
1.429061 |
AAAAAA |
AAAAAA |
| HNRNPAB |
1 |
351 |
0.004761905 |
0.0017734784 |
1.424957 |
AAAGAC |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPD |
7 |
1488 |
0.019047619 |
0.0075020153 |
1.344261 |
AAAAAA,AGAUAU,AUUUAU,UAUUUA,UUAGGA,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| PUM1 |
10 |
2172 |
0.026190476 |
0.0109482064 |
1.258348 |
GUAAAU,UAAAUA,UAAUGU,UACAUA,UAGAUA,UAUAUA,UGUAAA,UUAAUG |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| A1CF |
2 |
598 |
0.007142857 |
0.0030179363 |
1.242939 |
AUAAUU,UUAAUU |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPL |
6 |
1419 |
0.016666667 |
0.0071543732 |
1.220068 |
AAAUAA,AAUAAA,ACAUAC,CACAAA,CAUACA |
AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| TRA2B |
6 |
1447 |
0.016666667 |
0.0072954454 |
1.191898 |
AAGAAC,AAGGAA,AGAAGG,AGGAAA,GAAGGA |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| ELAVL1 |
15 |
3309 |
0.038095238 |
0.0166767432 |
1.191773 |
AUUUAU,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGUUUU,UUAUUU,UUGAUU,UUGUUU,UUUAGU,UUUAUU,UUUGUU,UUUUUG |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ELAVL3 |
8 |
1928 |
0.021428571 |
0.0097188634 |
1.140676 |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| RBM5 |
10 |
2359 |
0.026190476 |
0.0118903668 |
1.139249 |
AAAAAA,AAGGAA,AAGGAG,AGGGAA,CAAGGA,CUUCUC,GAAGGA,UCUUCU,UUCUCU |
AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| CELF1 |
4 |
1097 |
0.011904762 |
0.0055320435 |
1.105654 |
GUGUUG,GUUUGU,UGUGUU,UUGUGU |
CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CELF5 |
2 |
669 |
0.007142857 |
0.0033756550 |
1.081334 |
GUGUUG,UGUGUU |
GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| TRA2A |
4 |
1133 |
0.011904762 |
0.0057134220 |
1.059112 |
AAGAGG,AGAAAG,AGAGGA,GAGGAA |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| PABPC4 |
1 |
462 |
0.004761905 |
0.0023327287 |
1.029520 |
AAAAAA |
AAAAAA,AAAAAG |
| ELAVL2 |
41 |
9826 |
0.100000000 |
0.0495112858 |
1.014171 |
AAUUUG,AAUUUU,AUAUUU,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,GAUUUA,GAUUUU,GUUUUU,UAAUUU,UAUAUA,UAUGUU,UAUUUA,UAUUUU,UGAUUU,UUAAUU,UUACUU,UUAUUU,UUGAUU,UUUACU,UUUAGU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPA1 |
26 |
6340 |
0.064285714 |
0.0319478033 |
1.008781 |
AAAAAA,AAGAGG,AAGGAG,AGAGGA,AGAUAU,AGAUUA,AGGGUU,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAGGA,CCAAGG,GACUUA,GAGGAA,GCCAAG,GGACUU,GGGUUA,GUUUAG,UAAGUA,UAGGGU,UUAGAU,UUAUUU,UUUAUA,UUUAUU |
AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |