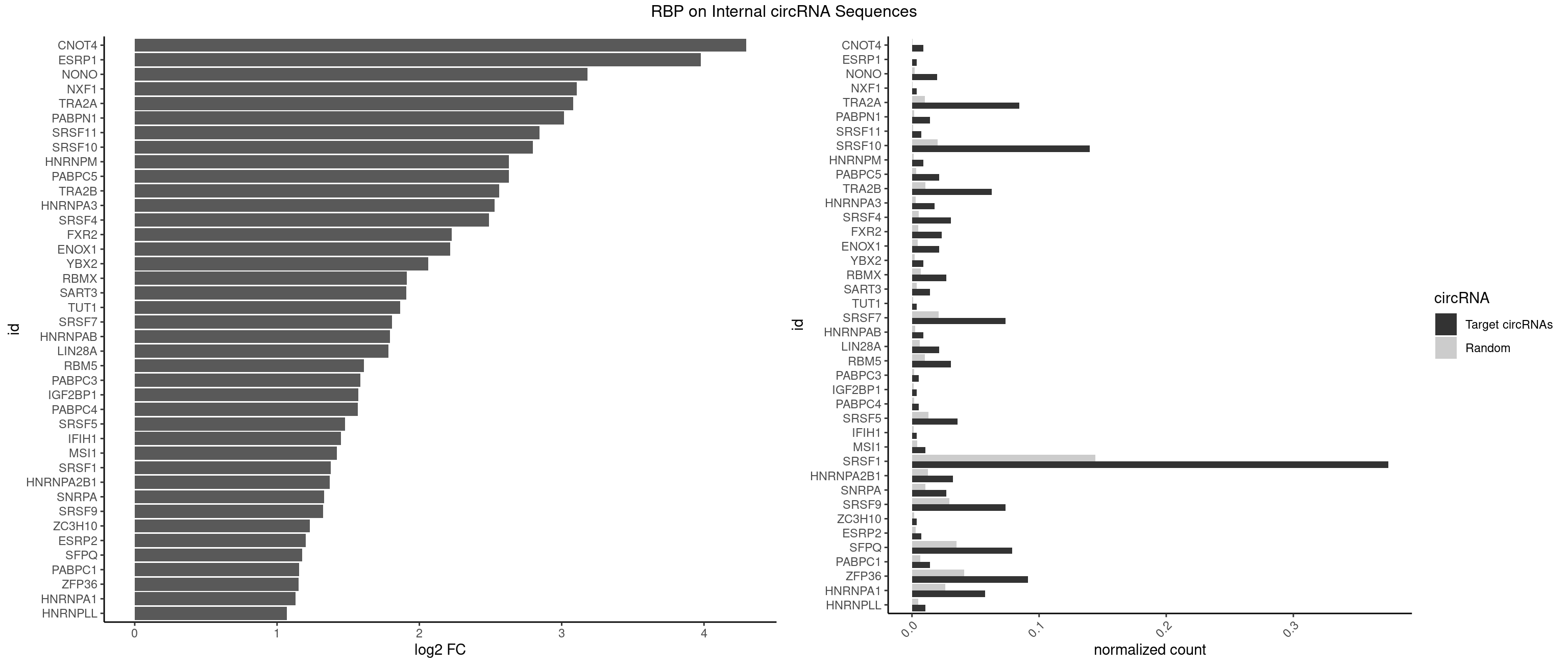
| CNOT4 |
4 |
314 |
0.008960573 |
0.0004564992 |
4.294907 |
GACAGA |
GACAGA |
| ESRP1 |
1 |
156 |
0.003584229 |
0.0002275250 |
3.977566 |
AGGGAU |
AGGGAU |
| NONO |
10 |
1498 |
0.019713262 |
0.0021723567 |
3.181834 |
AGAGGA,GAGAGG,GAGGAA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| NXF1 |
1 |
286 |
0.003584229 |
0.0004159215 |
3.107280 |
AACCUG |
AACCUG |
| TRA2A |
46 |
6871 |
0.084229391 |
0.0099589296 |
3.080261 |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| PABPN1 |
7 |
1222 |
0.014336918 |
0.0017723764 |
3.015978 |
AAAAGA,AGAAGA |
AAAAGA,AGAAGA |
| SRSF11 |
3 |
688 |
0.007168459 |
0.0009985015 |
2.843826 |
AAGAAG |
AAGAAG |
| SRSF10 |
77 |
13860 |
0.139784946 |
0.0200874160 |
2.798845 |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GAGAAA,GAGAAG,GAGACA,GAGACG,GAGAGA,GAGAGG,GAGGAA,GAGGAG,GAGGGA |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| HNRNPM |
4 |
999 |
0.008960573 |
0.0014492040 |
2.628330 |
AAGGAA,GAAGGA |
AAGGAA,GAAGGA,GGGGGG |
| PABPC5 |
11 |
2400 |
0.021505376 |
0.0034795387 |
2.627729 |
AGAAAA,AGAAAG |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TRA2B |
34 |
7329 |
0.062724014 |
0.0106226650 |
2.561872 |
AAAGAA,AAGAAC,AAGAAG,AAGGAA,AGAAGA,AGAAGG,AGGAAA,AGGAAG,GAAAGA,GAAGAA,GAAGGA,GGAAGG,UAAGAA |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| HNRNPA3 |
9 |
2140 |
0.017921147 |
0.0031027457 |
2.530046 |
AAGGAG,AGGAGC,CAAGGA,CCAAGG,GGAGCC |
AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| SRSF4 |
16 |
3740 |
0.030465950 |
0.0054214720 |
2.490441 |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| FXR2 |
12 |
3434 |
0.023297491 |
0.0049780156 |
2.226532 |
AGACAG,GACAGA,GACAGG,GGACAG |
AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| ENOX1 |
11 |
3195 |
0.021505376 |
0.0046316558 |
2.215097 |
AAGACA,AGACAG,AGGACA,AUACAG,CAGACA,CAUACA,GGACAG |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| YBX2 |
4 |
1480 |
0.008960573 |
0.0021462711 |
2.061759 |
AACAUC,ACAUCG,ACAUCU |
AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBMX |
14 |
4925 |
0.026881720 |
0.0071387787 |
1.912876 |
AAGAAG,AAGGAA,AAGUAA,ACCAAA,AGAAGG,AGGAAG,GAAGGA,GGAAGG |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SART3 |
7 |
2634 |
0.014336918 |
0.0038186524 |
1.908599 |
AGAAAA,GAAAAA,GAAAAC |
AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| TUT1 |
1 |
678 |
0.003584229 |
0.0009840095 |
1.864919 |
AGAUAC |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SRSF7 |
40 |
14481 |
0.073476703 |
0.0209873716 |
1.807765 |
AAAGGA,AAGAAG,AAGGAC,AGAAGA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGGAAG,AGGACA,CAGAGA,CCGAGA,CGAGAG,CUGAGA,GAAGAA,GAGAGA,GAGGAA,GAUAGA,GGAAGG,UAGAGA,UCGAGA |
AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| HNRNPAB |
4 |
1782 |
0.008960573 |
0.0025839306 |
1.794024 |
AAAGAC,AAGACA,CAAAGA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| LIN28A |
11 |
4315 |
0.021505376 |
0.0062547643 |
1.781670 |
AGGAGA,AGGAGU,CGGAGA,GGAGAA,GGAGAU |
AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| RBM5 |
16 |
6879 |
0.030465950 |
0.0099705232 |
1.611457 |
AAGGAA,AAGGAG,AGGGAA,AGGGAG,AGGGUG,CAAGGA,GAAGGA,GAAGGG,GAGGGA,GGUGGU |
AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| PABPC3 |
2 |
1234 |
0.005376344 |
0.0017897669 |
1.586854 |
AAAACA,GAAAAC |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| IGF2BP1 |
1 |
831 |
0.003584229 |
0.0012057377 |
1.571747 |
AAGCAC |
AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PABPC4 |
2 |
1251 |
0.005376344 |
0.0018144033 |
1.567130 |
AAAAAG |
AAAAAA,AAAAAG |
| SRSF5 |
19 |
8869 |
0.035842294 |
0.0128544391 |
1.479396 |
AAGAAG,AGAAGA,AGGAAG,CACAGA,CACAGC,GAAGAA,GAGGAA,GGAAGA,UGCGUA |
AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| IFIH1 |
1 |
904 |
0.003584229 |
0.0013115296 |
1.450413 |
GGCCGC |
CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| MSI1 |
5 |
2770 |
0.010752688 |
0.0040157442 |
1.420958 |
AGGAAG,AGGUGG,AGUAAG |
AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| SRSF1 |
208 |
99563 |
0.374551971 |
0.1442885423 |
1.376209 |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAGAAC,AAGAAG,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGAG,ACAGAA,ACAGAG,ACAGCA,ACAGGG,ACGCGA,AGAAGA,AGAAGG,AGACAG,AGAGAA,AGAGAC,AGAGGA,AGAGGG,AGAUGG,AGCCGA,AGCGGG,AGGAAA,AGGAAG,AGGACA,AGGAGA,AGGAGC,AUGAAG,AUGGAC,CAAGGA,CACAGA,CACAGC,CAGACA,CAGAGA,CAGAGG,CCAAGG,CCACCA,CCAGGA,CCCACC,CCCGAC,CCCGGA,CCGACC,CCGAGG,CCGCUA,CCGGCA,CCGGGA,CCGUGG,CCUGGA,CGACCA,CGACCG,CGCUCA,CGGAAG,CGGCAG,CGGGCG,CGUGGG,CUCCGG,CUCGUG,CUGAAC,GAAAGA,GAAAGG,GAACCA,GAAGAA,GAAGAC,GAAGAG,GAAGAU,GAAGCU,GAAGGA,GACAGA,GACAGG,GACGCA,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGCGG,GAGGAA,GAGGAG,GAGGCA,GAGGGA,GAUGAA,GAUGAC,GAUGAU,GAUGCC,GAUGGA,GCAGGG,GCCCAC,GCCCGG,GCGGAG,GCUCCG,GCUGCG,GCUGGG,GGAAAG,GGAAGA,GGAAGG,GGACAG,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCC,GGAGCG,GGCAGA,GGCCCA,GGCCCG,GGCCGA,GGCGGA,GGGCCA,GGGCGG,GGGGAG,GGUGCA,GGUGGC,GGUGGU,UCAGGU,UGAACA,UGAAGA,UGAAGG,UGGAAA,UGGAGC |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AACACG,AACAGC,AACCGG,AAGAAC,AAGAAG,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGAG,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACACGA,ACACGU,ACAGAA,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCCGG,ACCCGU,ACCGGA,ACCGGU,ACGAAC,ACGAAG,ACGAAU,ACGACG,ACGCGA,ACGCGC,ACGCUC,ACGGAA,ACGGAC,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGA,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCCGU,AGCGAG,AGCGGA,AGCGGG,AGCGGU,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGCGA,AGGUAA,AGUCGG,AUACGA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,AUUACG,AUUUCA,CAACCA,CAACCC,CAACCG,CAACGA,CAACGC,CAAGCA,CAAGGA,CAAUGG,CACACG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGA,CACCGG,CACGCA,CACGCU,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGCGG,CAGUCG,CAUGGU,CCAACC,CCAACG,CCAAGC,CCAAGG,CCAAUG,CCACAG,CCACCA,CCACCC,CCACCG,CCACGA,CCACGC,CCACGG,CCACUG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCAUGA,CCAUGG,CCCACC,CCCACG,CCCACU,CCCAGC,CCCAGG,CCCAUG,CCCCCA,CCCCCC,CCCCCG,CCCCGA,CCCCGC,CCCCGG,CCCGAC,CCCGCA,CCCGCC,CCCGCG,CCCGGA,CCCGGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCCUGA,CCCUGC,CCCUGG,CCGACA,CCGACC,CCGACG,CCGACU,CCGAGC,CCGAGG,CCGAUG,CCGCCA,CCGCCC,CCGCCG,CCGCGA,CCGCGC,CCGCGG,CCGCUA,CCGGAC,CCGGAG,CCGGCA,CCGGCC,CCGGCG,CCGGGA,CCGGGC,CCGGGG,CCGUCC,CCGUCG,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCA,CCUCCG,CCUCGA,CCUCGG,CCUGAC,CCUGCA,CCUGCG,CCUGGA,CCUGGG,CGAACC,CGAACG,CGAAGC,CGAAGG,CGACAG,CGACCA,CGACCC,CGACCG,CGACGA,CGACGC,CGACGG,CGAGCA,CGAGCG,CGAGGA,CGAGGC,CGAGGG,CGAUGA,CGAUGG,CGCACA,CGCACC,CGCACG,CGCAGC,CGCAGG,CGCCCA,CGCCCC,CGCCCG,CGCCGA,CGCCGC,CGCCGG,CGCCGU,CGCGAG,CGCGCA,CGCGCC,CGCGCG,CGCGCU,CGCGGA,CGCGGC,CGCGGG,CGCGGU,CGCGUC,CGCUAU,CGCUCA,CGCUCC,CGCUCG,CGCUGC,CGCUGG,CGGAAC,CGGAAG,CGGAAU,CGGACA,CGGACC,CGGACG,CGGAGC,CGGAGG,CGGCAC,CGGCAG,CGGCCA,CGGCCC,CGGCCG,CGGCGA,CGGCGC,CGGCGG,CGGGCA,CGGGCC,CGGGCG,CGGGGA,CGGGGC,CGGGGG,CGGUCC,CGGUCG,CGGUGC,CGGUGG,CGUCCA,CGUCCG,CGUCGA,CGUCGG,CGUGCA,CGUGCG,CGUGGA,CGUGGG,CUACGA,CUAGGG,CUCAGG,CUCCGG,CUCGGA,CUCGUG,CUGAAC,CUGACU,CUGAGC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAACCA,GAACGA,GAAGAA,GAAGAC,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACCCG,GACCGA,GACGAA,GACGAC,GACGAG,GACGCA,GACGCG,GACGGA,GACGGC,GACGGG,GACUGA,GACUGG,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGCGA,GAGCGG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUACG,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GAUGGU,GCACCA,GCACCG,GCACGA,GCACGG,GCACGU,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCAUAC,GCCCAC,GCCCCA,GCCCCG,GCCCGA,GCCCGG,GCCCGU,GCCGCA,GCCGCG,GCCGGA,GCCGGG,GCCGGU,GCCGUA,GCGAGC,GCGCAA,GCGCCA,GCGCCC,GCGCCG,GCGCGA,GCGCGC,GCGCGG,GCGGAA,GCGGAC,GCGGAG,GCGGCA,GCGGCG,GCGGGA,GCGGGG,GCGGUU,GCGUCA,GCUCCA,GCUCCG,GCUCGA,GCUCGG,GCUGCA,GCUGCG,GCUGGA,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGACGG,GGACGU,GGACUG,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCC,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAC,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCACG,GGCAGA,GGCCCA,GGCCCG,GGCCGA,GGCCGG,GGCCGU,GGCGCA,GGCGCG,GGCGGA,GGCGGG,GGCGGU,GGGACG,GGGCCA,GGGCCG,GGGCGA,GGGCGG,GGGGAA,GGGGAC,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGGGG,GGGUAC,GGUAAC,GGUCCA,GGUCCG,GGUCGA,GGUCGG,GGUGAA,GGUGAC,GGUGAU,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,GUGACG,UAAUUU,UACGAA,UACGGA,UAGACA,UAGGAC,UAUUAC,UCAAGA,UCAGGU,UCCGGA,UCGCAC,UCGGGC,UCGUGU,UGAACA,UGAACC,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUA,UGACUG,UGAGUU,UGAUGA,UGAUGC,UGAUGG,UGGAAA,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGA,UGGUGC,UGGUGG,UGUAGG,UUAAUU,UUACGG,UUCAAG,UUGACA,UUUCAA |
| HNRNPA2B1 |
17 |
8607 |
0.032258065 |
0.0124747476 |
1.370649 |
AAGAAG,AAGGAA,AAGGAG,AGAAGC,AGGAGC,CAAGGA,CCAAGG,GAAGGA,GGAGCC |
AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| SNRPA |
14 |
7380 |
0.026881720 |
0.0106965744 |
1.329477 |
ACCUGC,AGGAGA,AUACCU,CCUGCU,GAGCAG,GAUACC,GGAGAU,UACCUG,UUCCUG |
ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| SRSF9 |
40 |
20254 |
0.073476703 |
0.0293536261 |
1.323748 |
AGGAAA,AGGACA,AGGAGA,AGGAGC,AUGACC,GAAGGA,GAUGCC,GAUGGA,GGAAAA,GGAAAG,GGAAGG,GGACAG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAUGC,GGGUGG,GGUGCA,GGUGGC,UGAAAG,UGAACA,UGAAGG,UGGAGC |
AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| ZC3H10 |
1 |
1053 |
0.003584229 |
0.0015274610 |
1.230527 |
GGAGCG |
CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| ESRP2 |
3 |
2150 |
0.007168459 |
0.0031172377 |
1.201395 |
GGGAAA,GGGGAG,UGGGGA |
GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SFPQ |
43 |
24050 |
0.078853047 |
0.0348548043 |
1.177809 |
AAGAAC,AAGAGC,AAGAGG,AAGGAA,AAGGAC,AGAGAG,AGAGGA,AGGGAU,CUAAGG,CUGGAG,GAAGAA,GAAGAG,GAAGGA,GAGAGG,GAGGAA,GGAAGA,GGAGAG,GGGAUC,GUAAGA,GUGGUU,UAAGGA,UAGAGA,UGCAGG,UGGAGC |
AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
| PABPC1 |
7 |
4443 |
0.014336918 |
0.0064402624 |
1.154544 |
AGAAAA,GAAAAA,GAAAAC |
AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| ZFP36 |
50 |
28400 |
0.091397849 |
0.0411588414 |
1.150958 |
AAAAAG,AAAACA,AAAAGA,AAAAGC,AAAAGU,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAGAAA,AAGGAA,AAGUAA,ACCUGC,ACCUUC,AGAAAG,AGGAAA,AGUAAA,AUAAGG,CAAAGA,CAAGGA,CCUUCC,GGAAAG,GUAAAG,UAAGGA,UAAGUA |
AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
| HNRNPA1 |
31 |
18070 |
0.057347670 |
0.0261885646 |
1.130798 |
AAAGAG,AAGAGG,AAGGAG,AGAGGA,AGGAAG,AGGAGC,AGGGAC,AGUGAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAGGAA,GAGGAG,GGAAGG,GGAGCC,GUAAGU,UAAGUA,UAGAGA |
AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| HNRNPLL |
5 |
3534 |
0.010752688 |
0.0051229360 |
1.069655 |
ACCACA,CACACC,CACCAC,CACCGC,CAUACA |
ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |