circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000127663:+:19:5143966:5144902
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000127663:+:19:5143966:5144902 | ENSG00000127663 | ENST00000611640 | + | 19 | 5143967 | 5144902 | 471 | AAAUGCGUGUACUGCCGGAAGCGGAUGAAGAAGGUGUCAGGUGCCUGUAUCCAGUGCUCCUACGAGCACUGCUCCACGUCCUUCCACGUGACCUGCGCCCACGCCGCAGGCGUGCUCAUGGAGCCGGACGACUGGCCCUAUGUGGUCUCCAUCACCUGCCUCAAGCACAAGUCGGGGGGUCACGCUGUCCAACUCCUGAGGGCCGUGUCCCUAGGCCAGGUGGUCAUCACCAAGAACCGCAACGGGCUGUACUACCGCUGUCGCGUCAUCGGUGCCGCCUCGCAGACCUGCUACGAAGUGAACUUCGACGAUGGCUCCUACAGCGACAACCUGUACCCUGAGAGCAUCACGAGUAGGGACUGUGUCCAGCUGGGACCCCCUUCCGAGGGGGAGCUGGUGGAGCUCCGGUGGACUGACGGCAACCUCUACAAGGCCAAGUUCAUCUCCUCCGUCACCAGCCACAUCUACCAGAAAUGCGUGUACUGCCGGAAGCGGAUGAAGAAGGUGUCAGGUGCCUGUAU | circ |
| ENSG00000127663:+:19:5143966:5144902 | ENSG00000127663 | ENST00000611640 | + | 19 | 5143967 | 5144902 | 22 | ACAUCUACCAGAAAUGCGUGUA | bsj |
| ENSG00000127663:+:19:5143966:5144902 | ENSG00000127663 | ENST00000611640 | + | 19 | 5143767 | 5143976 | 210 | GGCUGGGUCAGUGGGUCUGGAGGGCUUGGCCAGGGAGGCUGCGGGCUCUGGGCUGUGGAGGGGAACCCUCACUGGGCAGAGCGCAGGGCCACUCCCGCGAUGCCUCCCUUGAAGGCUGUGCCGGGAGGGGCCGGGGACUCCGUUCCAGGGUCCCUAGGGAAGCUCGAGCCCCAUGCCCCUGCCUGUGUCCCCAUCCCCAGAAAUGCGUGU | ie_up |
| ENSG00000127663:+:19:5143966:5144902 | ENSG00000127663 | ENST00000611640 | + | 19 | 5144893 | 5145102 | 210 | CAUCUACCAGGUAAGCGGGGGAUCUGGCAGCCGCGCCAUGCCUUCACCAAGCUCUUCUUGUAGGUGCGGGGACAGGAGGAUCACACCCCUGGCCCAGGUGCCUUUGCCUGGGGCACUGGCGGGUGUGGGCCAUGGUUAGUGAGGCCCGCAGGACCCAGCUGAGCCUUGGCUCGCCUGCCUAAGUUAGAAGCACAGGGCUUGUUUGUUUUC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
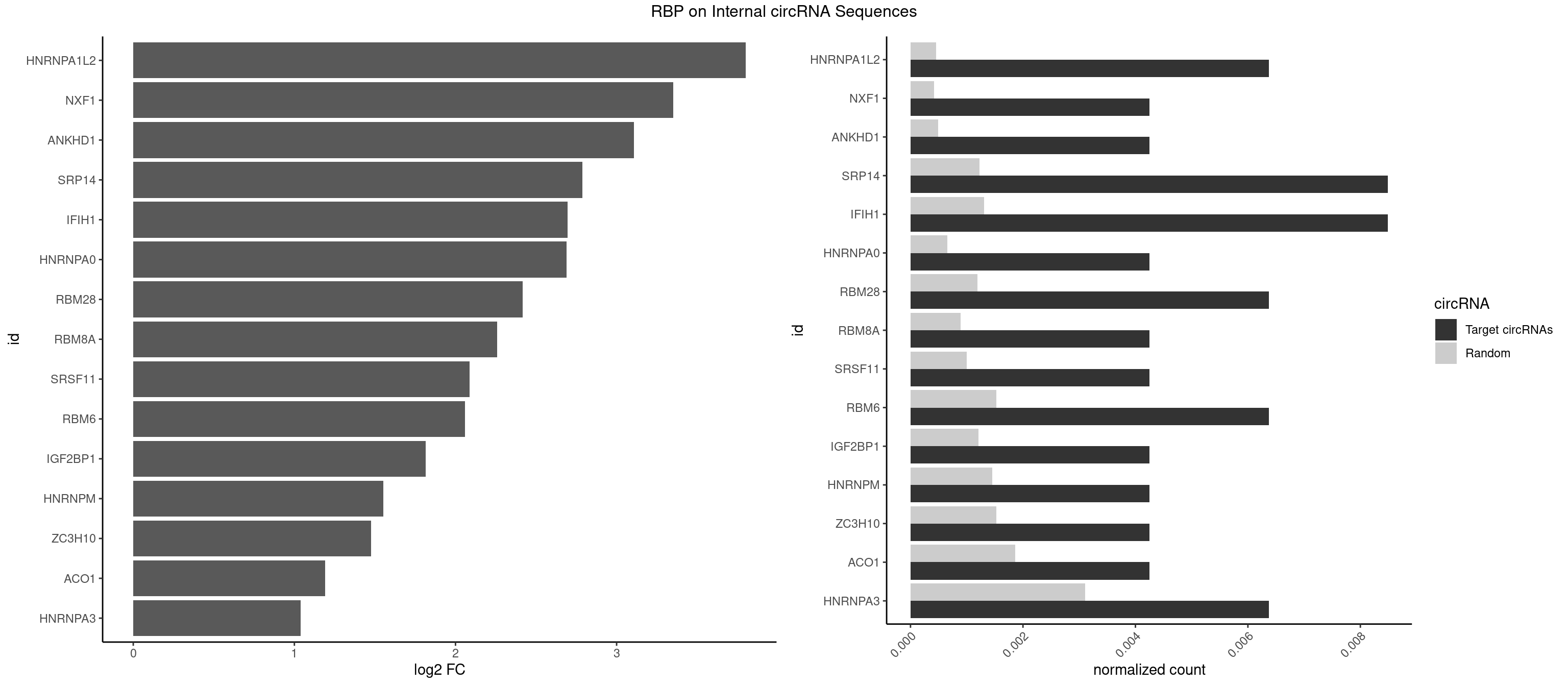
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 2 | 314 | 0.006369427 | 0.0004564992 | 3.802479 | GUAGGG,UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| NXF1 | 1 | 286 | 0.004246285 | 0.0004159215 | 3.351818 | AACCUG | AACCUG |
| ANKHD1 | 1 | 339 | 0.004246285 | 0.0004927293 | 3.107334 | GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SRP14 | 3 | 847 | 0.008492569 | 0.0012289250 | 2.788804 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 | 3 | 904 | 0.008492569 | 0.0013115296 | 2.694951 | GCGGAU,GGCCCU,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| HNRNPA0 | 1 | 453 | 0.004246285 | 0.0006579386 | 2.690176 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| RBM28 | 2 | 822 | 0.006369427 | 0.0011926949 | 2.416939 | AGUAGG,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBM8A | 1 | 611 | 0.004246285 | 0.0008869128 | 2.259337 | UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| SRSF11 | 1 | 688 | 0.004246285 | 0.0009985015 | 2.088365 | AAGAAG | AAGAAG |
| RBM6 | 2 | 1054 | 0.006369427 | 0.0015289102 | 2.058660 | AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| IGF2BP1 | 1 | 831 | 0.004246285 | 0.0012057377 | 1.816285 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPM | 1 | 999 | 0.004246285 | 0.0014492040 | 1.550940 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| ZC3H10 | 1 | 1053 | 0.004246285 | 0.0015274610 | 1.475066 | CAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| ACO1 | 1 | 1283 | 0.004246285 | 0.0018607779 | 1.190295 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPA3 | 2 | 2140 | 0.006369427 | 0.0031027457 | 1.037618 | GCCAAG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
RBP on BSJ (Exon Only)
Plot
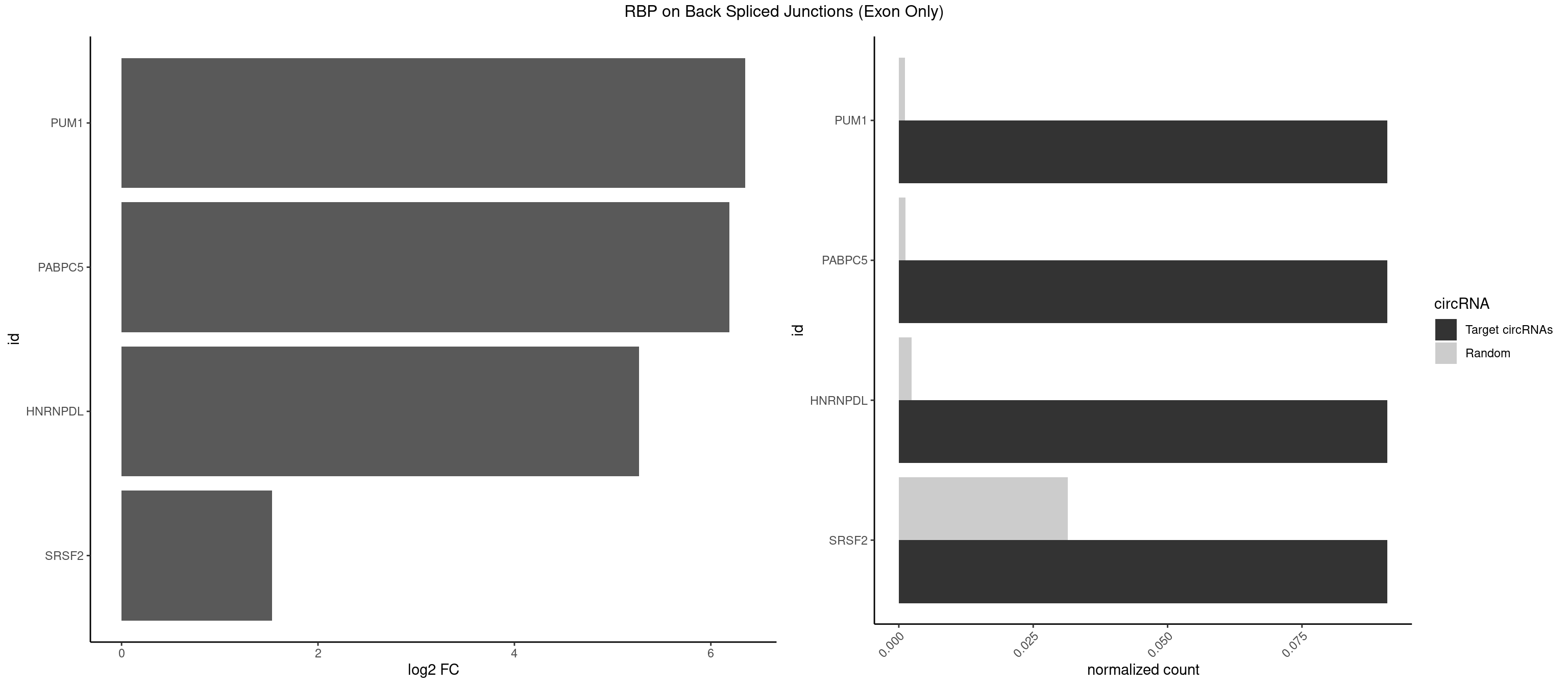
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PUM1 | 1 | 16 | 0.09090909 | 0.001113440 | 6.351329 | CCAGAA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAUAA,CAGAAU,GUAAAU,GUCCAG,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA,UUAAUG |
| PABPC5 | 1 | 18 | 0.09090909 | 0.001244433 | 6.190864 | AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPDL | 1 | 35 | 0.09090909 | 0.002357873 | 5.268867 | ACCAGA | AACAGC,AAUACC,AAUUUA,ACACCA,ACAGCA,ACCAGA,AGAUAU,AGUAGG,AUCUGA,AUUAGC,AUUAGG,CACGCA,CCAGAC,CUAAGC,CUAAGU,CUAGAU,CUAGGA,CUAGGC,CUUUAG,GAACUA,GAUUAG,GCACUA,GCUAGU,UGCGCA,UUAGCC,UUAGGC,UUUAGG |
| SRSF2 | 1 | 479 | 0.09090909 | 0.031438302 | 1.531901 | CCAGAA | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
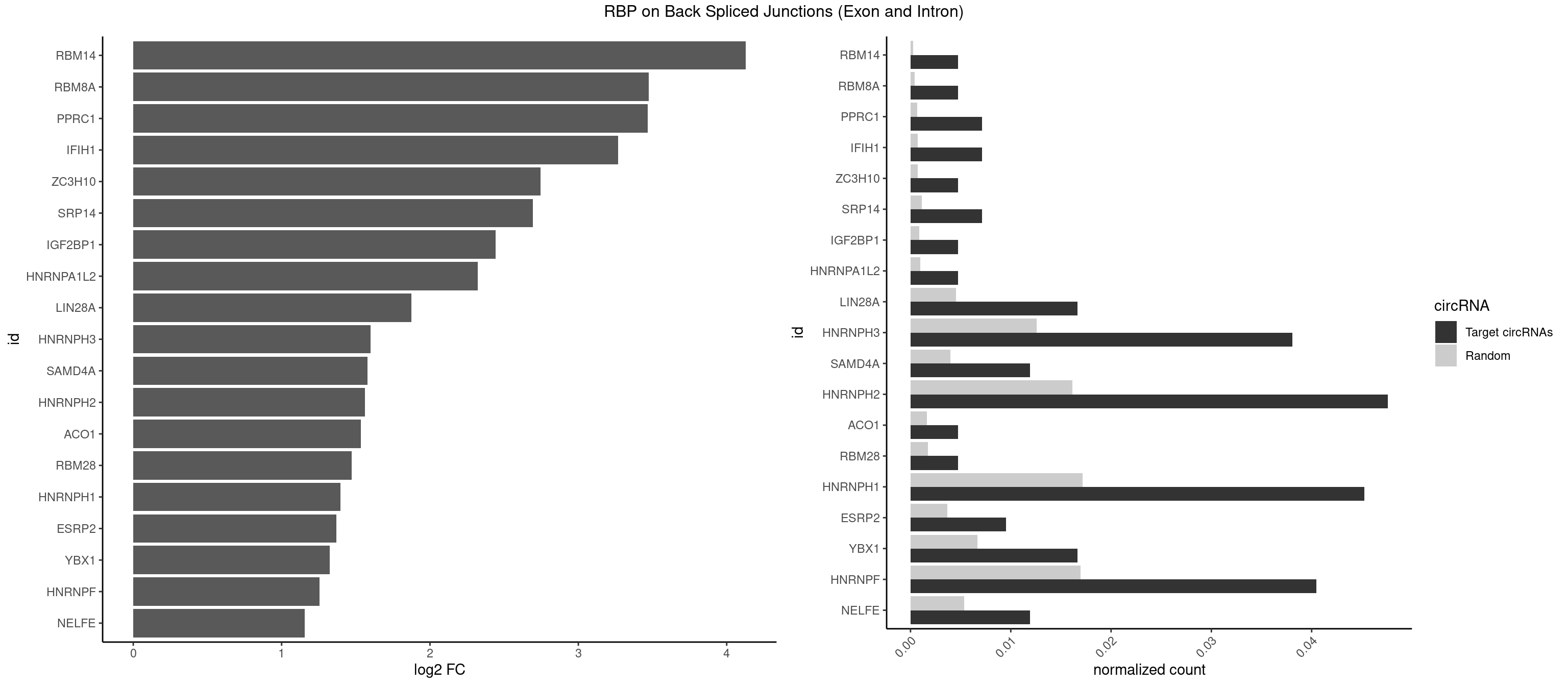
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM14 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | CGCGCC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBM8A | 1 | 84 | 0.004761905 | 0.0004282547 | 3.474998 | CGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| PPRC1 | 2 | 127 | 0.007142857 | 0.0006449012 | 3.469351 | CCGCGC,CGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| IFIH1 | 2 | 146 | 0.007142857 | 0.0007406288 | 3.269679 | GCCGCG,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | GAGCGC | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| LIN28A | 6 | 900 | 0.016666667 | 0.0045395002 | 1.876360 | GGAGGA,GGAGGG,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPH3 | 15 | 2496 | 0.038095238 | 0.0125806127 | 1.598408 | AGGGAA,AGGGGA,CGGGGG,GAGGGG,GGAGGA,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGUGU,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| SAMD4A | 4 | 790 | 0.011904762 | 0.0039852882 | 1.578783 | CUGGCA,CUGGCC,GCGGGC,GCGGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| HNRNPH2 | 19 | 3198 | 0.047619048 | 0.0161174929 | 1.562911 | AGGGAA,AGGGGA,CAGGAC,CGGGGG,CUGGGG,GAGGGG,GGAGGA,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGGAA,GGGGGA,GGGUGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPH1 | 18 | 3406 | 0.045238095 | 0.0171654575 | 1.398030 | AGGGAA,AGGGGA,CAGGAC,CGGGGG,CUGGGG,GAGGGG,GGAGGA,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGGAA,GGGUGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAG,GGGGAA,GGGGAU | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| YBX1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | CACACC,CCCUGC,CUGCGG,GAUCUG,GCCUGC,GGUCUG | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| HNRNPF | 16 | 3360 | 0.040476190 | 0.0169336961 | 1.257177 | AGGGAA,AGGGGA,CGGGGG,CUGGGG,GAGGGG,GGAGGA,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGGAA,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUCUGG,GGUUAG,UCUGGC,UGGUUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.