circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000079156:+:2:178284926:178285121
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000079156:+:2:178284926:178285121 | ENSG00000079156 | ENST00000190611 | + | 2 | 178284927 | 178285121 | 195 | GUUCUGCCACUUGCUGGGAAAACAGCAACAGAAGGAUAUUAAGGAGCCACCCCUAUAAUUUACCCAGAUAGCCCCAGCAAGGUCAAAGACAUAUCAUGUCCCAAGGAGAAAAGCUGCAGGCAUCUGAGGAUAAAUCACUGUGAAACAGCUUUGACCAUAAAGCUGACUUGGAAGACUUUGACUCCAAGGUGCAAGGUUCUGCCACUUGCUGGGAAAACAGCAACAGAAGGAUAUUAAGGAGCCAC | circ |
| ENSG00000079156:+:2:178284926:178285121 | ENSG00000079156 | ENST00000190611 | + | 2 | 178284927 | 178285121 | 22 | CAAGGUGCAAGGUUCUGCCACU | bsj |
| ENSG00000079156:+:2:178284926:178285121 | ENSG00000079156 | ENST00000190611 | + | 2 | 178284727 | 178284936 | 210 | UGAGAAGGUCAGUGGUUACGUCUUUGAGGUUACAUAAAACUGAACAAAGGAGAUAAAAAGCUGAGGCAUGUACCCAUAUGAACUUCCUGUUCAAAAGAUAAAUUGGGAGGAAUUUUCUGAUUGGCCAUAGGCAGUCCCCGUGAGAGGCUCACGAAAGUUUUGUAAAGAUGUACUAUAUGACUGUAAUCUCUCUUUUUCAGGUUCUGCCAC | ie_up |
| ENSG00000079156:+:2:178284926:178285121 | ENSG00000079156 | ENST00000190611 | + | 2 | 178285112 | 178285321 | 210 | AAGGUGCAAGGUGAGUUAGAAGAACACAAGCUUAAAACCAAAAGCAGGGUUAGGUUAGGCAGCGCUUAAUAAUAGCAACAGAUUAGUGGGACACCACAAAGUCACAGGAAGGUGUUUCUUUAUAUCUCUUAUUUGUAUGUCAGAUCUUAAAAUAAUAUGUCAUAAUCCUUAUAAUAAAAAGUUCUUUUUUUUUUAAGGAUAGGAAGUAGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
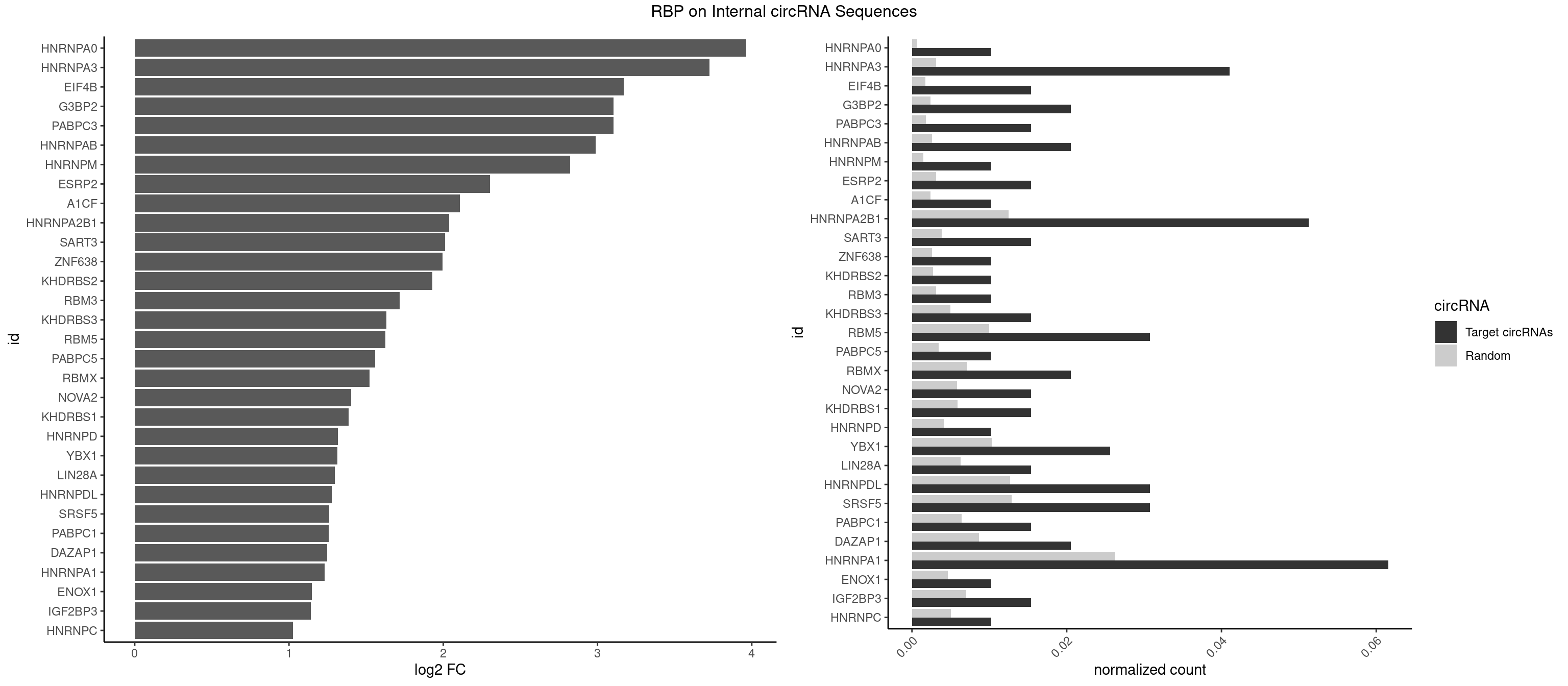
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 1 | 453 | 0.01025641 | 0.0006579386 | 3.962429 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| HNRNPA3 | 7 | 2140 | 0.04102564 | 0.0031027457 | 3.724909 | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| EIF4B | 2 | 1179 | 0.01538462 | 0.0017100607 | 3.169369 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| G3BP2 | 3 | 1644 | 0.02051282 | 0.0023839405 | 3.105106 | AGGAUA,GGAUAA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| PABPC3 | 2 | 1234 | 0.01538462 | 0.0017897669 | 3.103645 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPAB | 3 | 1782 | 0.02051282 | 0.0025839306 | 2.988887 | AAAGAC,AAGACA,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPM | 1 | 999 | 0.01025641 | 0.0014492040 | 2.823193 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ESRP2 | 2 | 2150 | 0.01538462 | 0.0031172377 | 2.303148 | GGGAAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| A1CF | 1 | 1642 | 0.01025641 | 0.0023810421 | 2.106861 | AUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPA2B1 | 9 | 8607 | 0.05128205 | 0.0124747476 | 2.039443 | AAGGAG,AAUUUA,AGGAGC,CAAGGA,CCAAGG,GAAGGA,GGAGCC | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| SART3 | 2 | 2634 | 0.01538462 | 0.0038186524 | 2.010353 | AGAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ZNF638 | 1 | 1773 | 0.01025641 | 0.0025708878 | 1.996187 | GGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| KHDRBS2 | 1 | 1858 | 0.01025641 | 0.0026940701 | 1.928667 | GAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RBM3 | 1 | 2152 | 0.01025641 | 0.0031201361 | 1.716845 | AAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| KHDRBS3 | 2 | 3429 | 0.01538462 | 0.0049707696 | 1.629947 | AUAAAG,GAUAAA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| RBM5 | 5 | 6879 | 0.03076923 | 0.0099705232 | 1.625747 | AAGGAG,AAGGUG,CAAGGA,GAAGGA | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| PABPC5 | 1 | 2400 | 0.01025641 | 0.0034795387 | 1.559558 | AGAAAA | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBMX | 3 | 4925 | 0.02051282 | 0.0071387787 | 1.522777 | AGAAGG,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| NOVA2 | 2 | 4013 | 0.01538462 | 0.0058171047 | 1.403115 | AGACAU,AGGCAU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS1 | 2 | 4064 | 0.01538462 | 0.0058910141 | 1.384900 | AUUUAC,GAAAAC | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| HNRNPD | 1 | 2837 | 0.01025641 | 0.0041128408 | 1.318319 | AAUUUA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| YBX1 | 4 | 7119 | 0.02564103 | 0.0103183321 | 1.313244 | CAGCAA,CAUCUG,CCAGCA | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| LIN28A | 2 | 4315 | 0.01538462 | 0.0062547643 | 1.298461 | AGGAGA,GGAGAA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPDL | 5 | 8759 | 0.03076923 | 0.0126950266 | 1.277225 | AACAGC,AAUUUA,ACAGCA,AUCUGA | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| SRSF5 | 5 | 8869 | 0.03076923 | 0.0128544391 | 1.259222 | AACAGC,AUAAAG,CAACAG,GGAAGA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| PABPC1 | 2 | 4443 | 0.01538462 | 0.0064402624 | 1.256297 | AGAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| DAZAP1 | 3 | 5964 | 0.02051282 | 0.0086445016 | 1.246671 | AAUUUA,AGGAUA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPA1 | 11 | 18070 | 0.06153846 | 0.0261885646 | 1.232551 | AAGGAG,AAGGUG,AAUUUA,AGGAGC,CAAAGA,CAAGGA,CCAAGG,GCAGGC,GGAGCC | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| ENOX1 | 1 | 3195 | 0.01025641 | 0.0046316558 | 1.146926 | AAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| IGF2BP3 | 2 | 4815 | 0.01538462 | 0.0069793662 | 1.140320 | AAAACA,AAAUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| HNRNPC | 1 | 3471 | 0.01025641 | 0.0050316361 | 1.027426 | GGAUAU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
RBP on BSJ (Exon Only)
Plot
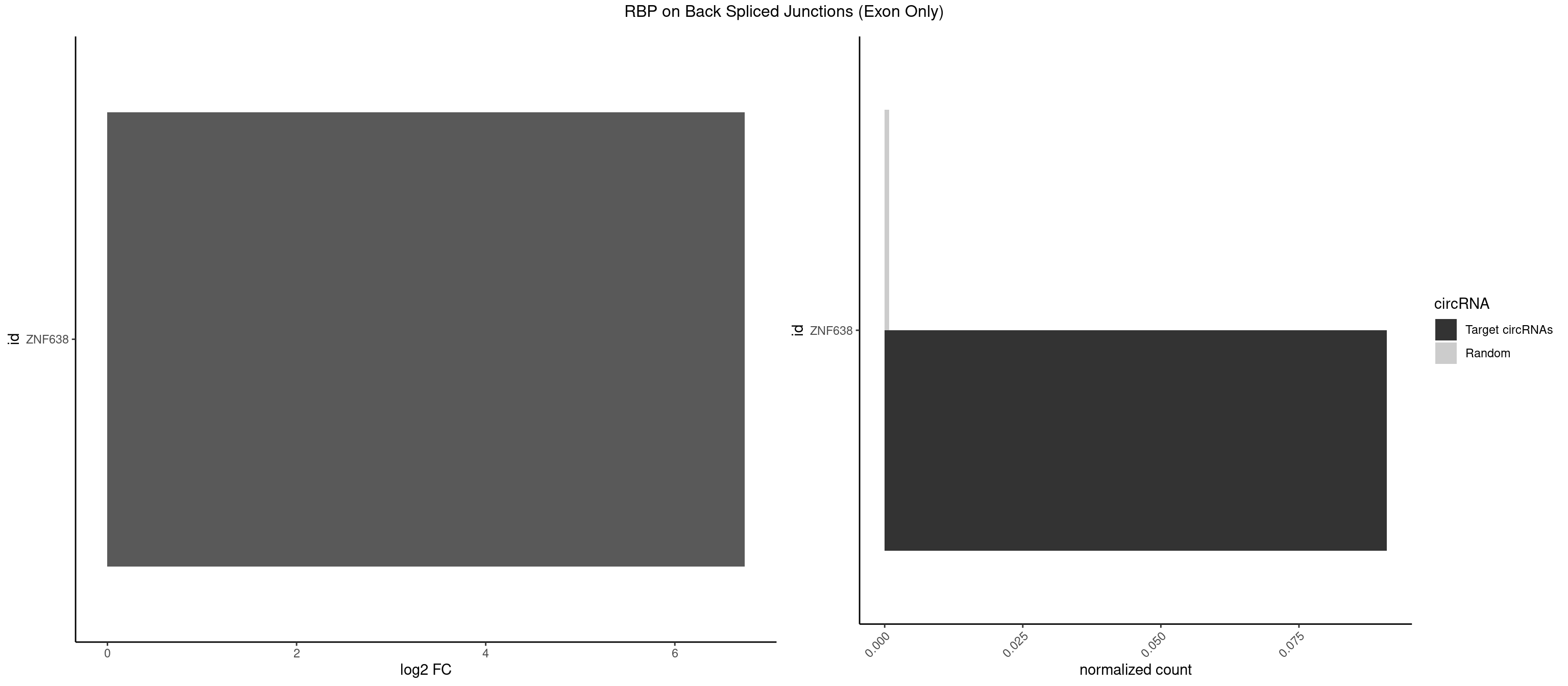
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZNF638 | 1 | 12 | 0.09090909 | 0.000851454 | 6.738352 | GGUUCU | CGUUCG,CGUUCU,CGUUGG,GGUUCU,GGUUGG,GUUCUU,GUUGGU,UGUUCU,UGUUGG,UGUUGU |
RBP on BSJ (Exon and Intron)
Plot
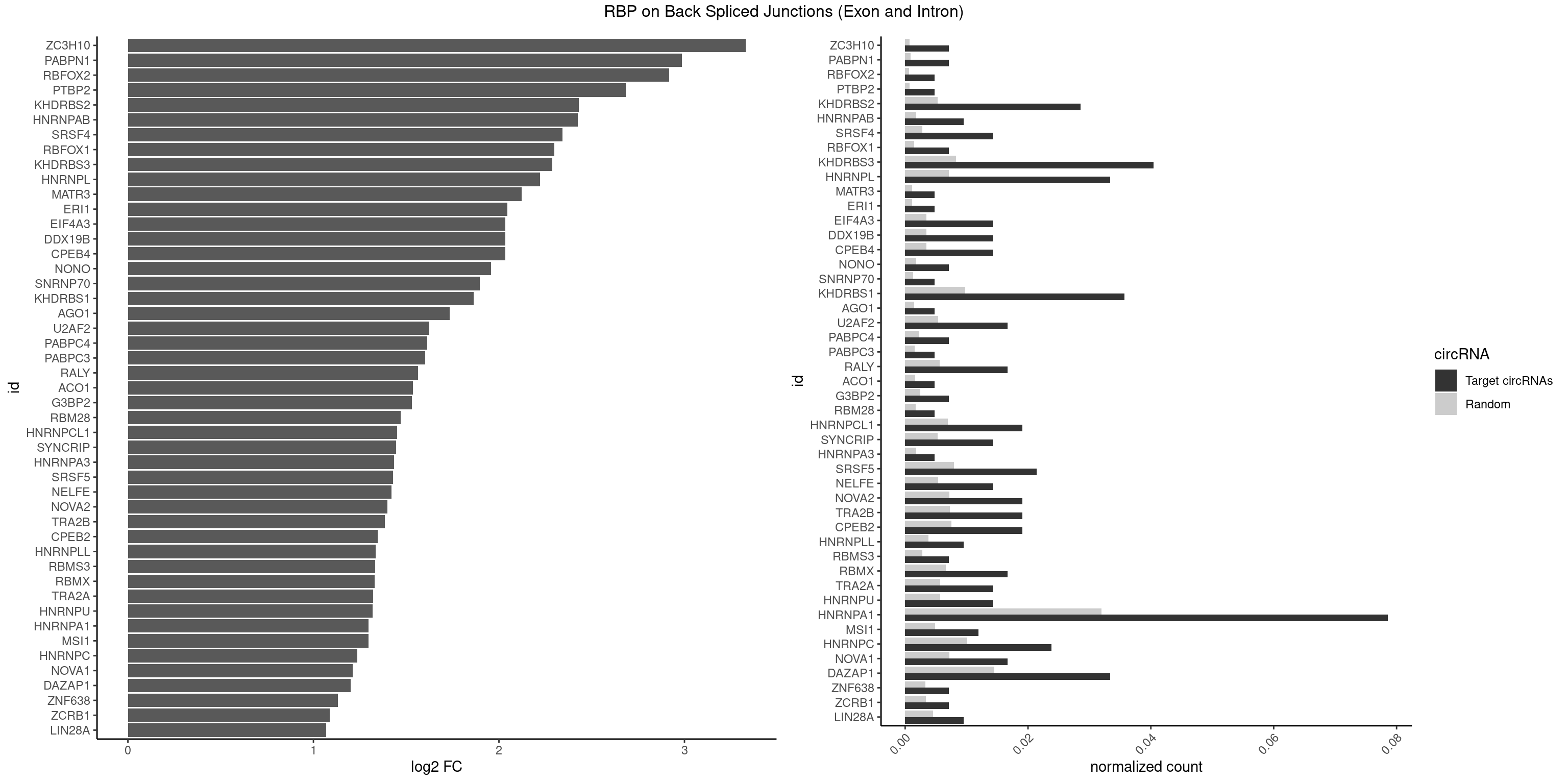
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZC3H10 | 2 | 140 | 0.007142857 | 0.0007103990 | 3.329800 | CAGCGC,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| KHDRBS2 | 11 | 1051 | 0.028571429 | 0.0053002821 | 2.430432 | AAUAAA,AUAAAA,GAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPAB | 3 | 351 | 0.009523810 | 0.0017734784 | 2.424957 | ACAAAG,AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SRSF4 | 5 | 558 | 0.014285714 | 0.0028164047 | 2.342647 | AGAAGA,AGGAAG,GAAGAA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGU,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| KHDRBS3 | 16 | 1646 | 0.040476190 | 0.0082980653 | 2.286227 | AAAUAA,AAUAAA,AGAUAA,AUAAAA,GAUAAA,UAAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPL | 13 | 1419 | 0.033333333 | 0.0071543732 | 2.220068 | AAAUAA,AACAAA,AACACA,AAUAAA,ACACAA,ACACCA,ACAUAA,ACCACA,CACAAA,CACAAG,CACCAC,CACGAA,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| CPEB4 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| DDX19B | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| NONO | 2 | 364 | 0.007142857 | 0.0018389762 | 1.957598 | GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| KHDRBS1 | 14 | 1945 | 0.035714286 | 0.0098045143 | 1.864983 | AUAAAA,UAAAAA,UAAAAC,UUAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| U2AF2 | 6 | 1071 | 0.016666667 | 0.0054010480 | 1.625654 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAG | AAAAAA,AAAAAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RALY | 6 | 1117 | 0.016666667 | 0.0056328094 | 1.565039 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUA,GGAUAG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPCL1 | 7 | 1381 | 0.019047619 | 0.0069629182 | 1.451847 | CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SYNCRIP | 5 | 1041 | 0.014285714 | 0.0052498992 | 1.444212 | UUUUUU | AAAAAA,UUUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| SRSF5 | 8 | 1578 | 0.021428571 | 0.0079554615 | 1.429518 | AGAAGA,AGGAAG,CAACAG,GAAGAA,GAGGAA,UACAUA,UACGUC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| NELFE | 5 | 1059 | 0.014285714 | 0.0053405885 | 1.419503 | CUCUCU,GGUUAG,UCUCUC,UGGUUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| NOVA2 | 7 | 1434 | 0.019047619 | 0.0072299476 | 1.397554 | AGGCAU,GAGGCA,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| TRA2B | 7 | 1447 | 0.019047619 | 0.0072954454 | 1.384543 | AAGAAC,AGAAGA,AGAAGG,AGGAAG,GAAGAA,GGAAGG | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| CPEB2 | 7 | 1487 | 0.019047619 | 0.0074969770 | 1.345230 | CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPLL | 3 | 748 | 0.009523810 | 0.0037736800 | 1.335567 | ACACCA,ACCACA,CACCAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | CUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBMX | 6 | 1316 | 0.016666667 | 0.0066354293 | 1.328704 | ACCAAA,AGAAGG,AGGAAG,GGAAGG,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| TRA2A | 5 | 1133 | 0.014285714 | 0.0057134220 | 1.322146 | AGAAGA,AGGAAG,GAAGAA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| HNRNPU | 5 | 1136 | 0.014285714 | 0.0057285369 | 1.318335 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPA1 | 32 | 6340 | 0.078571429 | 0.0319478033 | 1.298288 | AAGGAG,AAGGUG,AGAUUA,AGGAAG,AGGGUU,AGUGGU,AGUUAG,AUAGCA,GAAGGU,GAGGAA,GAUUAG,GGAAGG,GGAGGA,GGGUUA,GGUUAG,GUUAGG,UAGGAA,UAGGCA,UUAUUU,UUUAUA,UUUUUU | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGAAG,AGUUAG,UAGGAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| HNRNPC | 9 | 2006 | 0.023809524 | 0.0101118501 | 1.235492 | CUUUUU,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| NOVA1 | 6 | 1428 | 0.016666667 | 0.0071997179 | 1.210953 | AGUCAC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| DAZAP1 | 13 | 2878 | 0.033333333 | 0.0145052398 | 1.200391 | AGGAAG,AGGAUA,AGGUUA,AGUUAG,UAGGAA,UAGGUU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU,GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GCUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | AGGAGA,GGAGAU,GGAGGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.