circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000105281:-:19:46776974:46784559
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000105281:-:19:46776974:46784559 | ENSG00000105281 | ENST00000542575 | - | 19 | 46776975 | 46784559 | 822 | AAAUAUCUUCCCUUCCAACCUGGUGUCAGCAGCCUUUCGCUCAUACUCUACCACCUAUGAAGAGAGGAAUAUCACCGGAACCAGGGUGAAGGUGCCCGUGGGGCAGGAGGUGGAGGGGAUGAACAUCCUGGGCUUGGUAGUGUUUGCCAUCGUCUUUGGUGUGGCGCUGCGGAAGCUGGGGCCUGAAGGGGAGCUGCUUAUCCGCUUCUUCAACUCCUUCAAUGAGGCCACCAUGGUUCUGGUCUCCUGGAUCAUGUGGUACGCCCCUGUGGGCAUCAUGUUCCUGGUGGCUGGCAAGAUCGUGGAGAUGGAGGAUGUGGGUUUACUCUUUGCCCGCCUUGGCAAGUACAUUCUGUGCUGCCUGCUGGGUCACGCCAUCCAUGGGCUCCUGGUACUGCCCCUCAUCUACUUCCUCUUCACCCGCAAAAACCCCUACCGCUUCCUGUGGGGCAUCGUGACGCCGCUGGCCACUGCCUUUGGGACCUCUUCCAGUUCCGCCACGCUGCCGCUGAUGAUGAAGUGCGUGGAGGAGAAUAAUGGCGUGGCCAAGCACAUCAGCCGUUUCAUCCUGCCCAUCGGCGCCACCGUCAACAUGGACGGUGCCGCGCUCUUCCAGUGCGUGGCCGCAGUGUUCAUUGCACAGCUCAGCCAGCAGUCCUUGGACUUCGUAAAGAUCAUCACCAUCCUGGUCACGGCCACAGCGUCCAGCGUGGGGGCAGCGGGCAUCCCUGCUGGAGGUGUCCUCACUCUGGCCAUCAUCCUCGAAGCAGUCAACCUCCCGGUCGACCAUAUCUCCUUGAUCCUGGCUGUGGACUGGCUAGUAAAUAUCUUCCCUUCCAACCUGGUGUCAGCAGCCUUUCGCUCAUACUCUA | circ |
| ENSG00000105281:-:19:46776974:46784559 | ENSG00000105281 | ENST00000542575 | - | 19 | 46776975 | 46784559 | 22 | GACUGGCUAGUAAAUAUCUUCC | bsj |
| ENSG00000105281:-:19:46776974:46784559 | ENSG00000105281 | ENST00000542575 | - | 19 | 46784550 | 46784759 | 210 | GGCUGACUCCCCAAGUCCUGCAAAGGGGUGCAAGCCUAGUUCCAACCCUGCCGGGCUGAGGCAUGAGUAGGGGCUGUAUUUGCGUGCAGGCGGGCCCCCAAAGCUUCCACUCGCUGCCUUAAAACGCCAACCCAGCCUCUCAGACAAUGCUGCCCUCCCACUAUGGUAUCUAAUAACCCUCUGUGUGUCUGUCUCUGCAGAAAUAUCUUC | ie_up |
| ENSG00000105281:-:19:46776974:46784559 | ENSG00000105281 | ENST00000542575 | - | 19 | 46776775 | 46776984 | 210 | ACUGGCUAGUGUGAGUGUGGGUCUGGAGACUGGGGCAGGGCCAGGGGUACCCCUGGGAAAGAGGAUGAGUGAGUGAGAGUCCUUAGAGCCUCUGGACAGAAGAGGAAGAGAAGAAGGAAGGAAUUCUGGAGGAAAUGGGGGGAACCUUGAGCAUUCCUGAGGGCAAGGAAUGGCAUCUUUGUGGCUCUAAGAGGGAUGUCAGAAAAGUAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
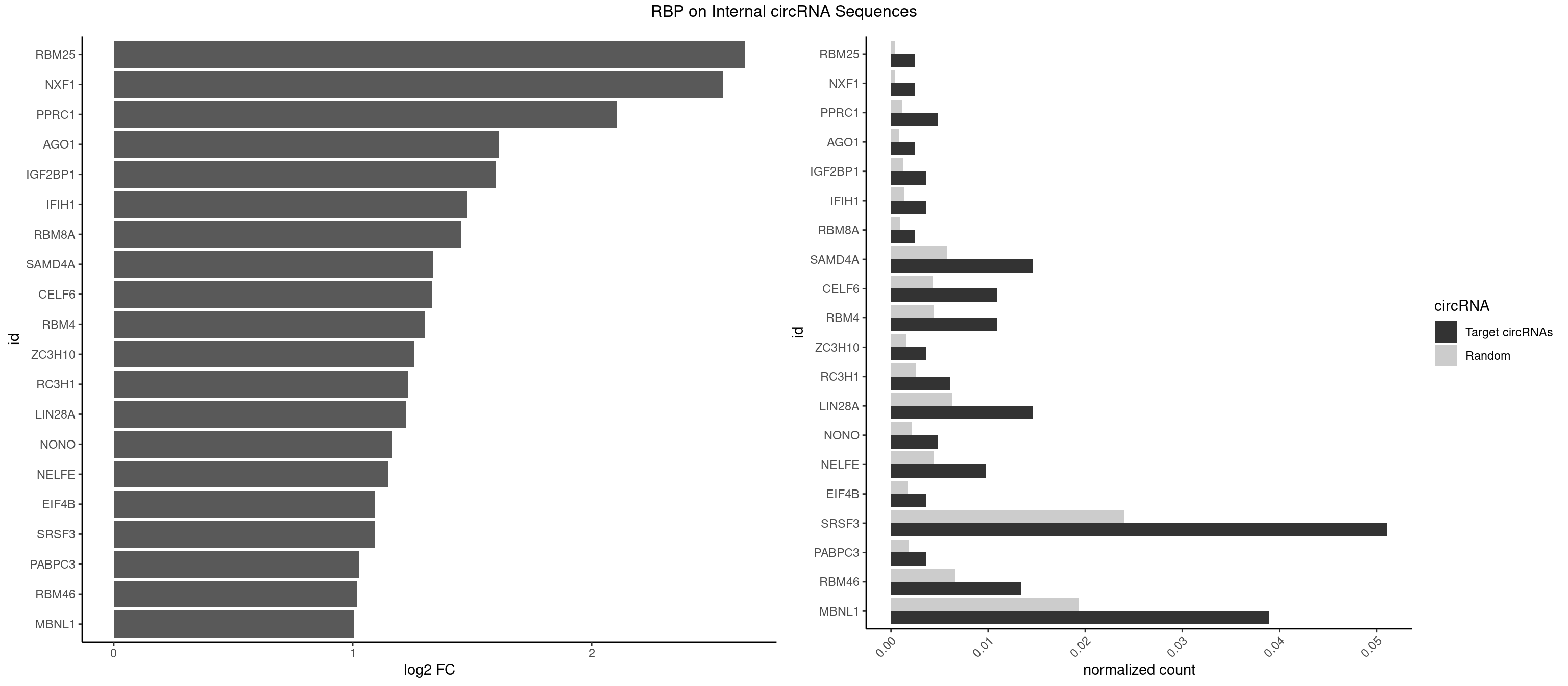
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 268 | 0.002433090 | 0.0003898359 | 2.641851 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| NXF1 | 1 | 286 | 0.002433090 | 0.0004159215 | 2.548406 | AACCUG | AACCUG |
| PPRC1 | 3 | 780 | 0.004866180 | 0.0011318283 | 2.104135 | CCGCGC,CGGCGC,GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| AGO1 | 1 | 548 | 0.002433090 | 0.0007956130 | 1.612651 | GGUAGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| IGF2BP1 | 2 | 831 | 0.003649635 | 0.0012057377 | 1.597836 | AAGCAC,CACCCG | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| IFIH1 | 2 | 904 | 0.003649635 | 0.0013115296 | 1.476502 | GCCGCG,GGCCGC | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM8A | 1 | 611 | 0.002433090 | 0.0008869128 | 1.455925 | CGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| SAMD4A | 11 | 3992 | 0.014598540 | 0.0057866714 | 1.335018 | CGGGCA,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGC,GCUGGA,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| CELF6 | 8 | 2997 | 0.010948905 | 0.0043447134 | 1.333454 | GUGGGG,GUGUGG,UGUGGG,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM4 | 8 | 3065 | 0.010948905 | 0.0044432593 | 1.301096 | CCUCUU,CCUUCC,CUCUUU,CUUCCU,CUUCUU,UCCUUC | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ZC3H10 | 2 | 1053 | 0.003649635 | 0.0015274610 | 1.256617 | CCAGCG,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RC3H1 | 4 | 1786 | 0.006082725 | 0.0025897275 | 1.231918 | CCCUUC,UCCCUU,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| LIN28A | 11 | 4315 | 0.014598540 | 0.0062547643 | 1.222797 | AGGAGA,GGAGAA,GGAGAU,GGAGGA,GGAGGG,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| NONO | 3 | 1498 | 0.004866180 | 0.0021723567 | 1.163529 | AGAGGA,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| NELFE | 7 | 3028 | 0.009732360 | 0.0043896388 | 1.148687 | CUCUGG,CUGGCU,GGUCUC,UCUGGC,UCUGGU,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| EIF4B | 2 | 1179 | 0.003649635 | 0.0017100607 | 1.093705 | CUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SRSF3 | 41 | 16536 | 0.051094891 | 0.0239654858 | 1.092221 | ACAUCA,ACAUUC,ACCACC,ACUUCG,AUCAUC,AUCUUC,CACAUC,CAUCAC,CAUCAU,CAUCGU,CCACCA,CCUCUU,CGCUUC,CUCUUC,CUUCAA,CUUCAC,CUUCUU,GACUUC,GUCAAC,UACUUC,UCAUCA,UCAUCC,UCAUCU,UCUUCA,UCUUCC,UUCAAC,UUCAUC,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| PABPC3 | 2 | 1234 | 0.003649635 | 0.0017897669 | 1.027981 | AAAAAC,AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RBM46 | 10 | 4554 | 0.013381995 | 0.0066011240 | 1.019510 | AUCAUG,AUGAAG,AUGAUG,GAUCAU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| MBNL1 | 31 | 13372 | 0.038929440 | 0.0193802045 | 1.006278 | ACGCUG,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUUC,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUG,CUGCUU,GCGCUC,GCGCUG,GCUGCG,GCUGCU,GUGCUG,UCCGCU,UCGCUC,UGCUGC,UUCGCU,UUGCCA,UUGCCC | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
RBP on BSJ (Exon Only)
Plot
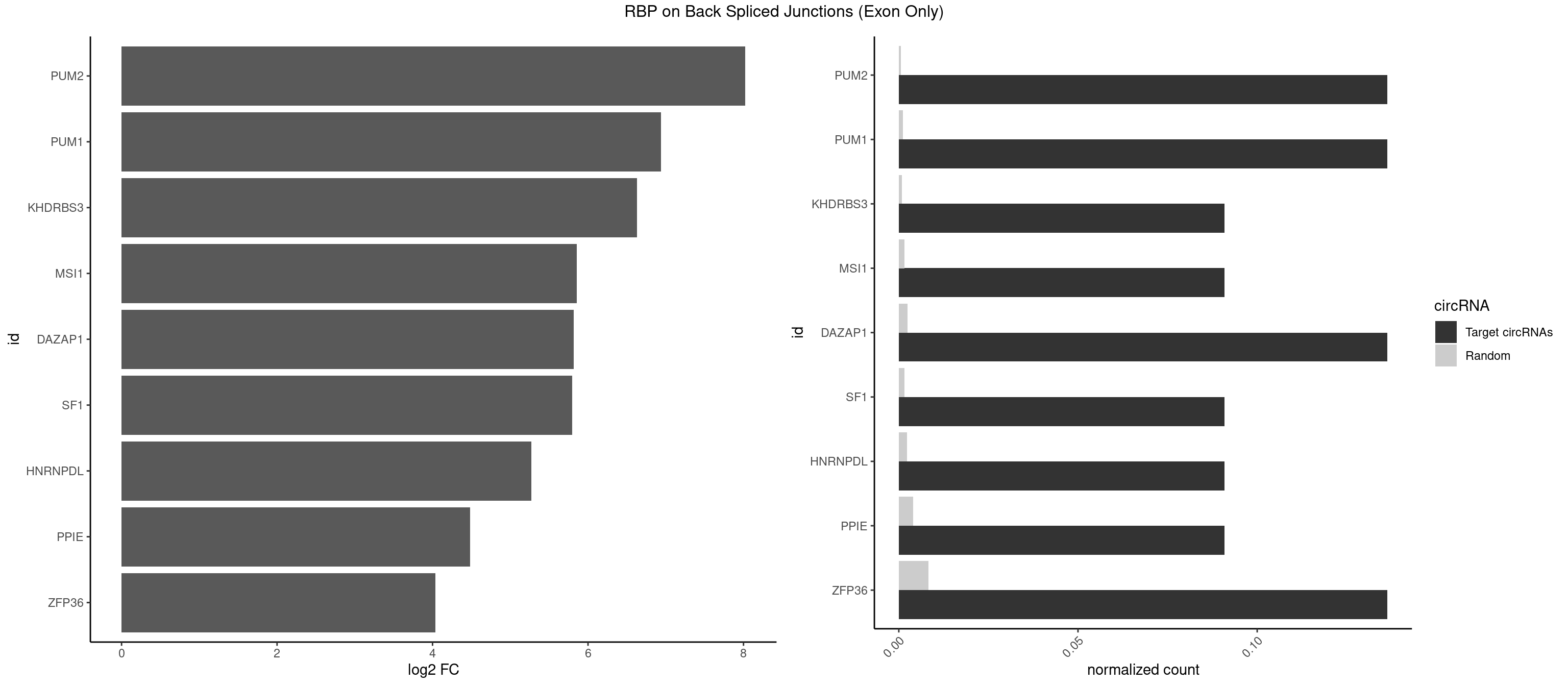
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PUM2 | 2 | 7 | 0.13636364 | 0.0005239717 | 8.023754 | GUAAAU,UAAAUA | GUAAAU,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA |
| PUM1 | 2 | 16 | 0.13636364 | 0.0011134399 | 6.936292 | GUAAAU,UAAAUA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAUAA,CAGAAU,GUAAAU,GUCCAG,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA,UUAAUG |
| KHDRBS3 | 1 | 13 | 0.09090909 | 0.0009169505 | 6.631437 | UAAAUA | AAAUAA,AGAUAA,AUAAAG,AUUAAA,GAUAAA,UAAAUA,UUAAAC |
| MSI1 | 1 | 23 | 0.09090909 | 0.0015719151 | 5.853829 | UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUGG,UAGGAA,UAGGAG,UAGGUG,UAGUUA,UAGUUG |
| DAZAP1 | 2 | 36 | 0.13636364 | 0.0024233691 | 5.814301 | AGUAAA,UAGUAA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| SF1 | 1 | 24 | 0.09090909 | 0.0016374116 | 5.794936 | UAGUAA | ACAGAC,ACAGUC,AGUAAG,AUACUA,CACAGA,CACUGA,CAGUCA,GCUGAC,GCUGCC,UACUGA,UGCUAA,UGCUGA,UGCUGC |
| HNRNPDL | 1 | 35 | 0.09090909 | 0.0023578727 | 5.268867 | CUAGUA | AACAGC,AAUACC,AAUUUA,ACACCA,ACAGCA,ACCAGA,AGAUAU,AGUAGG,AUCUGA,AUUAGC,AUUAGG,CACGCA,CCAGAC,CUAAGC,CUAAGU,CUAGAU,CUAGGA,CUAGGC,CUUUAG,GAACUA,GAUUAG,GCACUA,GCUAGU,UGCGCA,UUAGCC,UUAGGC,UUUAGG |
| PPIE | 1 | 61 | 0.09090909 | 0.0040607807 | 4.484596 | UAAAUA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAU,AUAAUA,AUAUAA,AUAUAU,AUAUUU,AUUAAA,AUUUAA,UAAAAA,UAAAUA,UAAAUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUU,UUAAAA,UUAAAU,UUAUUA |
| ZFP36 | 2 | 126 | 0.13636364 | 0.0083180508 | 4.035070 | AGUAAA,UAAAUA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
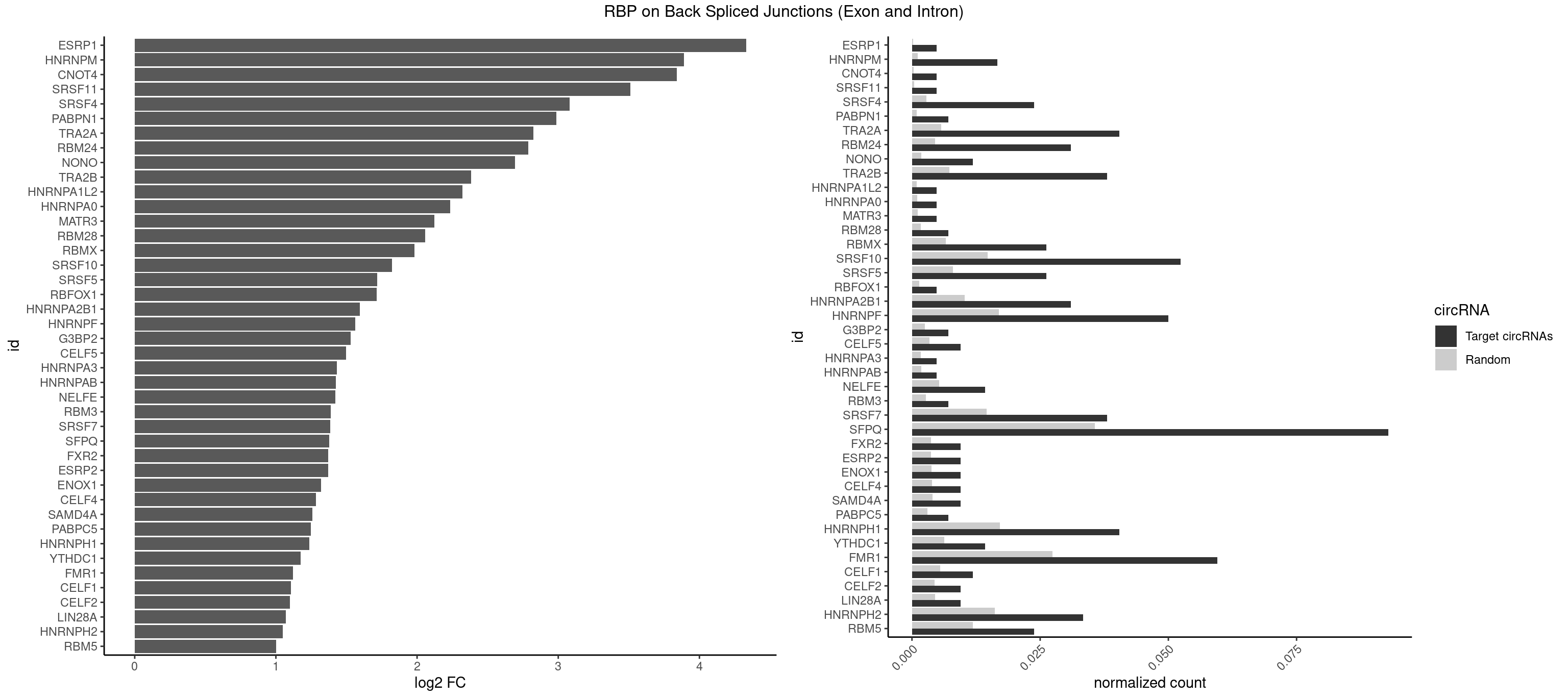
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 46 | 0.004761905 | 0.0002367997 | 4.329800 | AGGGAU | AGGGAU |
| HNRNPM | 6 | 222 | 0.016666667 | 0.0011235389 | 3.890844 | AAGGAA,GAAGGA,GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| SRSF4 | 9 | 558 | 0.023809524 | 0.0028164047 | 3.079612 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AGAAGA | AAAAGA,AGAAGA |
| TRA2A | 16 | 1133 | 0.040476190 | 0.0057134220 | 2.824647 | AAGAAG,AAGAGG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RBM24 | 12 | 888 | 0.030952381 | 0.0044790407 | 2.788789 | AGUGUG,GAGUGA,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| NONO | 4 | 364 | 0.011904762 | 0.0018389762 | 2.694564 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| TRA2B | 15 | 1447 | 0.038095238 | 0.0072954454 | 2.384543 | AAGAAG,AAGGAA,AGAAGA,AGAAGG,AGGAAA,AGGAAG,GAAAGA,GAAGAA,GAAGGA,GGAAGG | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | GUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGG,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBMX | 10 | 1316 | 0.026190476 | 0.0066354293 | 1.980781 | AAGAAG,AAGGAA,AGAAGG,AGGAAG,GAAGGA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SRSF10 | 21 | 2937 | 0.052380952 | 0.0148024990 | 1.823202 | AAAGAG,AAAGGG,AAGAAG,AAGAGA,AAGAGG,AAGGAA,AAGGGG,AGACAA,AGAGAA,AGAGGA,AGAGGG,GAAAGA,GAGAAG,GAGGAA,GAGGGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SRSF5 | 10 | 1578 | 0.026190476 | 0.0079554615 | 1.719025 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA,UGCAGA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA2B1 | 12 | 2033 | 0.030952381 | 0.0102478839 | 1.594724 | AAGAAG,AAGGAA,AAGGGG,AGGGGC,AGUAGG,CAAGGA,GAAGGA,GGAACC,GUAGGG | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| HNRNPF | 20 | 3360 | 0.050000000 | 0.0169336961 | 1.562031 | AAGGGG,AGGAAG,AGGGAU,AUGGGG,CUGGGG,GAGGAA,GGAAGG,GGAAUG,GGAGGA,GGGAUG,GGGCUG,GGGGAA,GGGGCU,GGGGGG,GGGGUA,UGGGAA,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGG,GUGUGU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NELFE | 5 | 1059 | 0.014285714 | 0.0053405885 | 1.419503 | CUCUGG,CUGGCU,GUCUCU,UCUCUG,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACG,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SRSF7 | 15 | 2893 | 0.038095238 | 0.0145808142 | 1.385539 | AAGAAG,AGAAGA,AGAGAA,AGAGGA,AGGAAG,GAAGAA,GAGACU,GAGGAA,GGAAGG,UGAGAG,UGGACA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| SFPQ | 38 | 7087 | 0.092857143 | 0.0357114067 | 1.378628 | AAGAGG,AAGGAA,ACUGGG,AGAGGA,AGGGAU,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGGA,GACUGG,GAGGAA,GCAGGC,GGAAGA,GGAGGA,GGGGGA,GGGGGG,GGUCUG,GUCUGG,UAAGAG,UAGGGG,UAGUGU,UCUAAG,UCUGGA,UGCAGG,UGGAGA,UGGAGG | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
| FXR2 | 3 | 730 | 0.009523810 | 0.0036829907 | 1.370661 | AGACAA,GACAGA,GGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGGAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | CAGACA,GGACAG,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| CELF4 | 3 | 776 | 0.009523810 | 0.0039147521 | 1.282618 | GUGUGG,GUGUGU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CGGGCC,CUGGAC,GCGGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPH1 | 16 | 3406 | 0.040476190 | 0.0171654575 | 1.237565 | AAGGGG,AGGAAG,CUGGGG,GAGGAA,GGAAGG,GGAAUG,GGAGGA,GGGCUG,GGGGAA,GGGGCU,GGGGGG,UGGGGG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GCGUGC,GGGUAC,GGGUGC,UCCUGC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| FMR1 | 24 | 5430 | 0.059523810 | 0.0273629585 | 1.121243 | AAGGAA,ACUGGG,AGGAAG,AGGAAU,AGGAUG,CUAUGG,CUGAGG,GAAGGA,GACUGG,GAGGAU,GCUGAG,GGACAG,GGCUGA,GGGCUG,GGGGGG,GUGGCU | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | CUGUCU,GUGUGU,UGUCUG,UGUGUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUCUGU,GUGUGU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | GGAGGA,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPH2 | 13 | 3198 | 0.033333333 | 0.0161174929 | 1.048338 | AAGGGG,CUGGGG,GGAAGG,GGAAUG,GGAGGA,GGGCUG,GGGGAA,GGGGCU,GGGGGA,GGGGGG,UGGGGG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| RBM5 | 9 | 2359 | 0.023809524 | 0.0118903668 | 1.001746 | AAGGAA,AAGGGG,CAAGGA,GAAGGA,GAGGGA,GGGGGG | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.