circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000181090_ENSG00000197070:+:9:137754170:137800984
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000181090_ENSG00000197070:+:9:137754170:137800984 | ENSG00000181090_ENSG00000197070 | MSTRG.33733.4 | + | 9 | 137754171 | 137800984 | 1464 | AGUUCGGAAUCCAGCAUUAAGAAGAAAUUUCUCAAGAGGAAAGGAAAGACCGACAGUCCCUGGAUCAAGCCAGCCAGGAAAAGGAGGCGGAGAAGUAGAAAGAAGCCCAGCGGUGCCCUCGGUUCUGAGUCGUAUAAGUCAUCUGCAGGAAGCGCUGAGCAGACGGCACCAGGAGACAGCACAGGGUACAUGGAAGUUUCUCUGGACUCCCUGGAUCUCCGAGUCAAAGGAAUUCUGUCUUCACAAGCAGAAGGGUUGGCCAACGGUCCAGAUGUGCUGGAGACAGACGGCCUCCAGGAAGUGCCUCUCUGCAGCUGCCGGAUGGAAACACCGAAGAGUCGAGAGAUCACCACACUGGCCAACAACCAGUGCAUGGCUACAGAGAGCGUGGACCAUGAAUUGGGCCGGUGCACAAACAGCGUGGUCAAGUAUGAGCUGAUGCGCCCCUCCAACAAGGCCCCGCUCCUCGUGCUGUGUGAAGACCACCGGGGCCGCAUGGUGAAGCACCAGUGCUGUCCUGGCUGUGGCUACUUCUGCACAGCGGGUAAUUUUAUGGAGUGUCAGCCCGAGAGCAGCAUCUCUCACCGUUUCCACAAAGACUGUGCCUCUCGAGUCAAUAACGCCAGCUAUUGUCCCCACUGUGGGGAGGAGAGCUCCAAGGCCAAAGAGGUGACGAUAGCUAAAGCAGACACCACCUCGACCGUGACACCAGUCCCCGGGCAGGAGAAGGGCUCGGCCCUGGAGGGCAGGGCCGACACCACAACGGGCAGUGCUGCCGGGCCACCACUCUCGGAGGACGACAAGCUGCAGGGUGCAGCCUCCCACGUGCCCGAGGGCUUUGAUCCAACGGGACCUGCUGGGCUUGGGAGGCCAACUCCCGGCCUUUCCCAGGGACCAGGGAAGGAAACCUUGGAGAGCGCUCUCAUCGCCCUCGACUCGGAAAAACCCAAGAAGCUUCGCUUCCACCCAAAGCAGCUGUACUUCUCCGCCAGGCAAGGGGAGCUUCAGAAGGUGCUCCUCAUGCUGGUGGACGGAAUUGACCCCAACUUCAAAAUGGAGCACCAGAAUAAGCGCUCUCCACUGCACGCCGCGGCAGAGGCUGGACACGUGGACAUCUGCCACAUGCUGGUUCAGGCGGGCGCUAAUAUUGACACCUGCUCAGAAGACCAGAGGACCCCGUUGAUGGAAGCAGCCGAAAACAACCAUCUGGAAGCAGUGAAGUACCUCAUCAAGGCUGGGGCCCUGGUGGAUCCCAAGGACGCAGAGGGCUCUACGUGUUUGCACCUGGCUGCCAAGAAAGGCCACUACGAAGUGGUCCAGUACCUGCUUUCAAAUGGACAGAUGGACGUCAACUGUCAGGAUGACGGAGGCUGGACACCCAUGAUCUGGGCCACAGAGUACAAGCACGUGGACCUCGUGAAGCUGCUGCUGUCCAAGGGCUCUGACAUCAACAUCCGAGACAACAGUUCGGAAUCCAGCAUUAAGAAGAAAUUUCUCAAGAGGAAAGGAAAGAC | circ |
| ENSG00000181090_ENSG00000197070:+:9:137754170:137800984 | ENSG00000181090_ENSG00000197070 | MSTRG.33733.4 | + | 9 | 137754171 | 137800984 | 22 | UCCGAGACAACAGUUCGGAAUC | bsj |
| ENSG00000181090_ENSG00000197070:+:9:137754170:137800984 | ENSG00000181090_ENSG00000197070 | MSTRG.33733.4 | + | 9 | 137753971 | 137754180 | 210 | UGAGCUGAGAGAGUUAGGGAAAAGCGCAGGGUGCCUUUAAGCGUGUGACCCAGCAAAAUGUCUACAAGUUAAGUUCACCGUUUAAUUGAAGAUCAAGACAUAGCCAGUGUUCUGUUUUGCAAGAAAACACUUGUAGUGCUCUUGCCUUCCGUAGCUUCCUGUGCUUAGUGGUUUAUAUGCCUGCCCGUUUGGUUCUCCAGAGUUCGGAAU | ie_up |
| ENSG00000181090_ENSG00000197070:+:9:137754170:137800984 | ENSG00000181090_ENSG00000197070 | MSTRG.33733.4 | + | 9 | 137800975 | 137801184 | 210 | CCGAGACAACGUAAGUUCGUCACACCCUCCCCGGGAGCCGUGUCCUGGAGGGGUGGGGACCUCCUCCCCCAGAAGGUUCUUUUCUCAAAAGCAGAACCCUGGCACCUGGUGGCGGCUUUGACCUCUUCCUGUGGCAGCAUCGGCCCGGCACUGUGCUGUGGCUGUCCUGUGCUGGAUGCAGAUGGGGGAUGUGAGGUCCCAGCAGAGGCC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
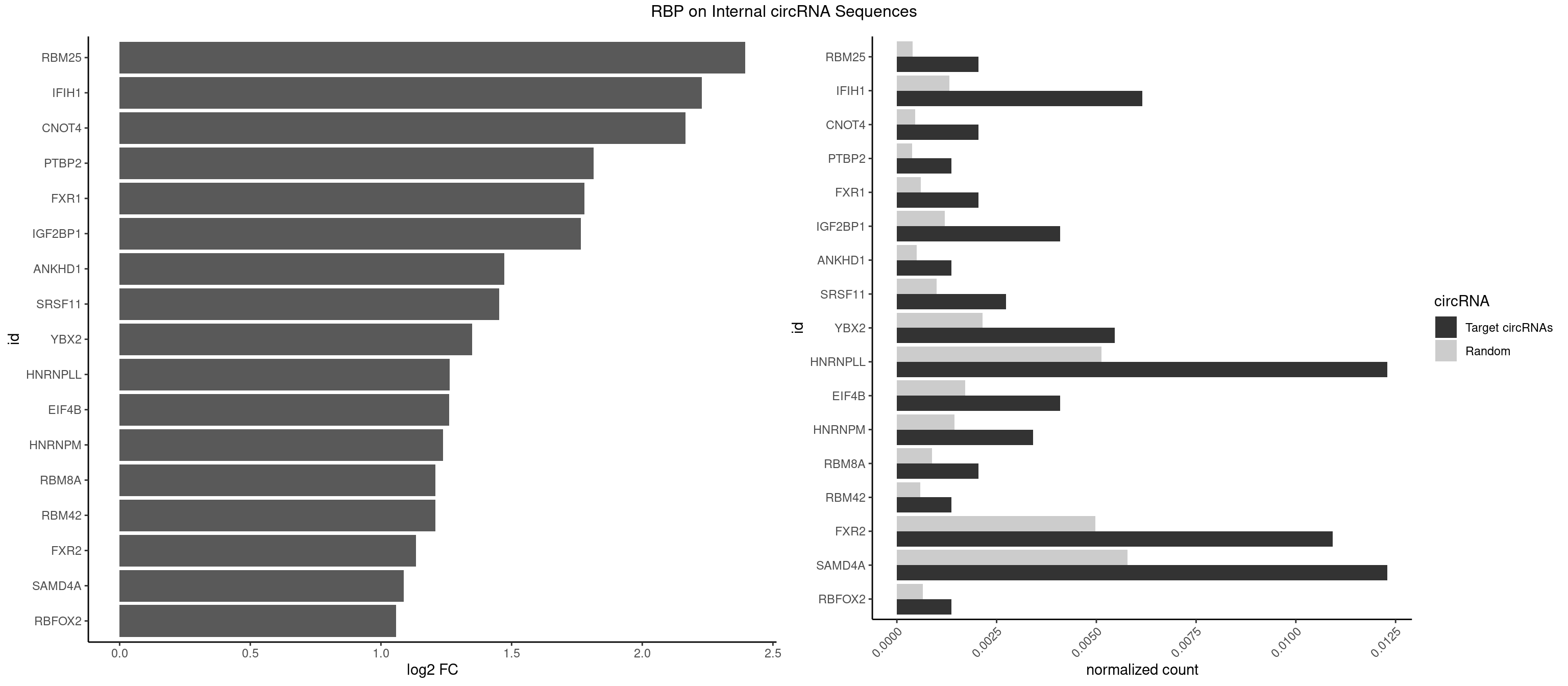
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 2 | 268 | 0.002049180 | 0.0003898359 | 2.394108 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| IFIH1 | 8 | 904 | 0.006147541 | 0.0013115296 | 2.228759 | CCGCGG,GCCGCG,GGCCCU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| CNOT4 | 2 | 314 | 0.002049180 | 0.0004564992 | 2.166363 | GACAGA | GACAGA |
| PTBP2 | 1 | 267 | 0.001366120 | 0.0003883867 | 1.814519 | CUCUCU | CUCUCU |
| FXR1 | 2 | 411 | 0.002049180 | 0.0005970720 | 1.779070 | ACGACA,AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| IGF2BP1 | 5 | 831 | 0.004098361 | 0.0012057377 | 1.765131 | AAGCAC,AGCACC,CCCGUU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ANKHD1 | 1 | 339 | 0.001366120 | 0.0004927293 | 1.471217 | GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SRSF11 | 3 | 688 | 0.002732240 | 0.0009985015 | 1.452248 | AAGAAG | AAGAAG |
| YBX2 | 7 | 1480 | 0.005464481 | 0.0021462711 | 1.348252 | AACAAC,AACAUC,ACAACA,ACAACG,ACAUCA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| HNRNPLL | 17 | 3534 | 0.012295082 | 0.0051229360 | 1.263039 | ACAAAC,ACACCA,ACCACA,ACGACA,ACUGCA,CAAACA,CACCAC,CACUGC,CAGACG,GACGAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| EIF4B | 5 | 1179 | 0.004098361 | 0.0017100607 | 1.260999 | CUCGGA,CUUGGA,UCGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPM | 4 | 999 | 0.003415301 | 0.0014492040 | 1.236752 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM8A | 2 | 611 | 0.002049180 | 0.0008869128 | 1.208183 | AUGCGC,UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| RBM42 | 1 | 407 | 0.001366120 | 0.0005912752 | 1.208183 | ACUACG | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR2 | 15 | 3434 | 0.010928962 | 0.0049780156 | 1.134514 | AGACAA,AGACAG,AGACGG,GACAAG,GACAGA,GACGGA,GGACAG,GGACGA,GGACGG,UGACGA,UGACGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| SAMD4A | 17 | 3992 | 0.012295082 | 0.0057866714 | 1.087276 | CGGGAC,CGGGCA,CGGGCC,CGGGUA,CUGGAA,CUGGAC,CUGGCC,GCGGGC,GCGGGU,GCUGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| RBFOX2 | 1 | 452 | 0.001366120 | 0.0006564894 | 1.057241 | UGCAUG | UGACUG,UGCAUG |
RBP on BSJ (Exon Only)
Plot
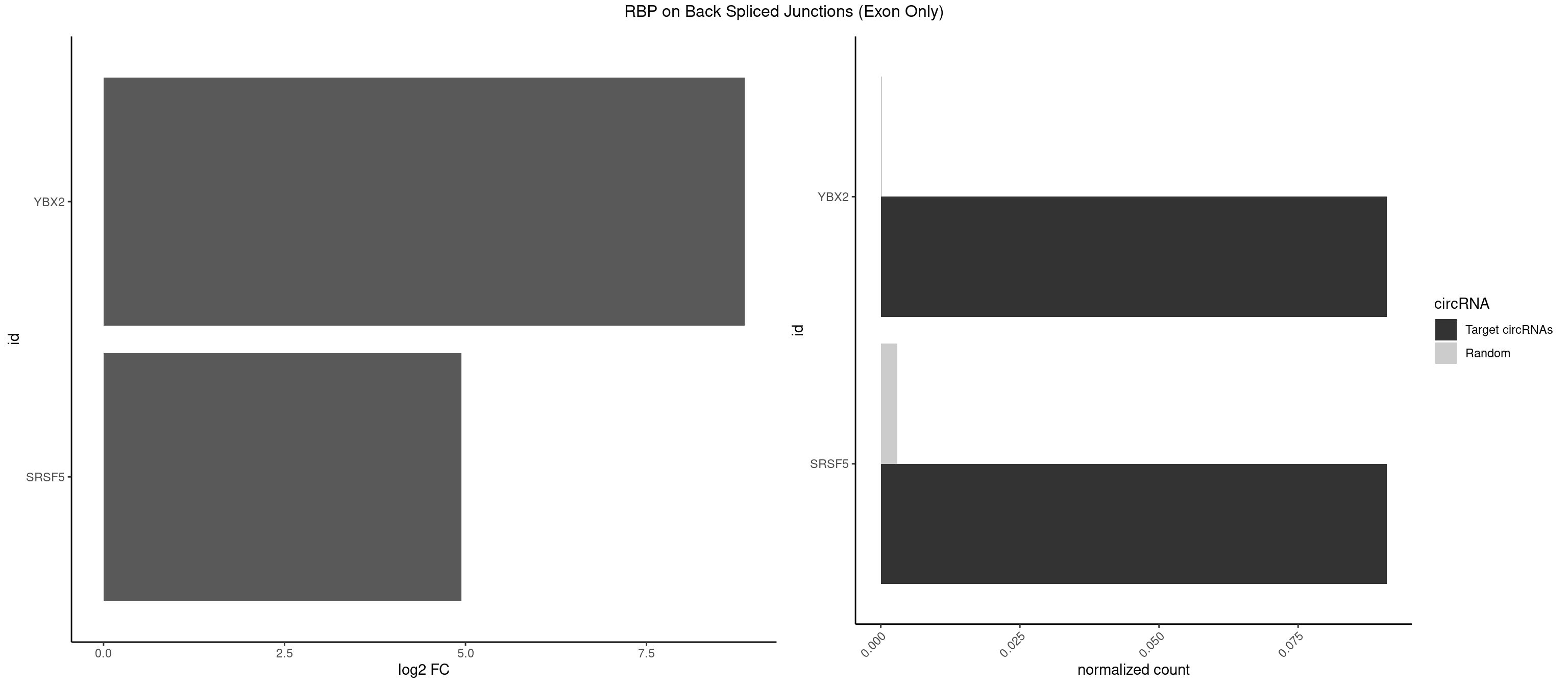
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| YBX2 | 1 | 2 | 0.09090909 | 0.0001964894 | 8.853829 | ACAACA | ACAACA,ACAUCU |
| SRSF5 | 1 | 44 | 0.09090909 | 0.0029473408 | 4.946939 | CAACAG | AACAGC,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUC,CACGGA,CGCAGC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACGUA,UGCAGA,UGCAGC,UGCGGC |
RBP on BSJ (Exon and Intron)
Plot
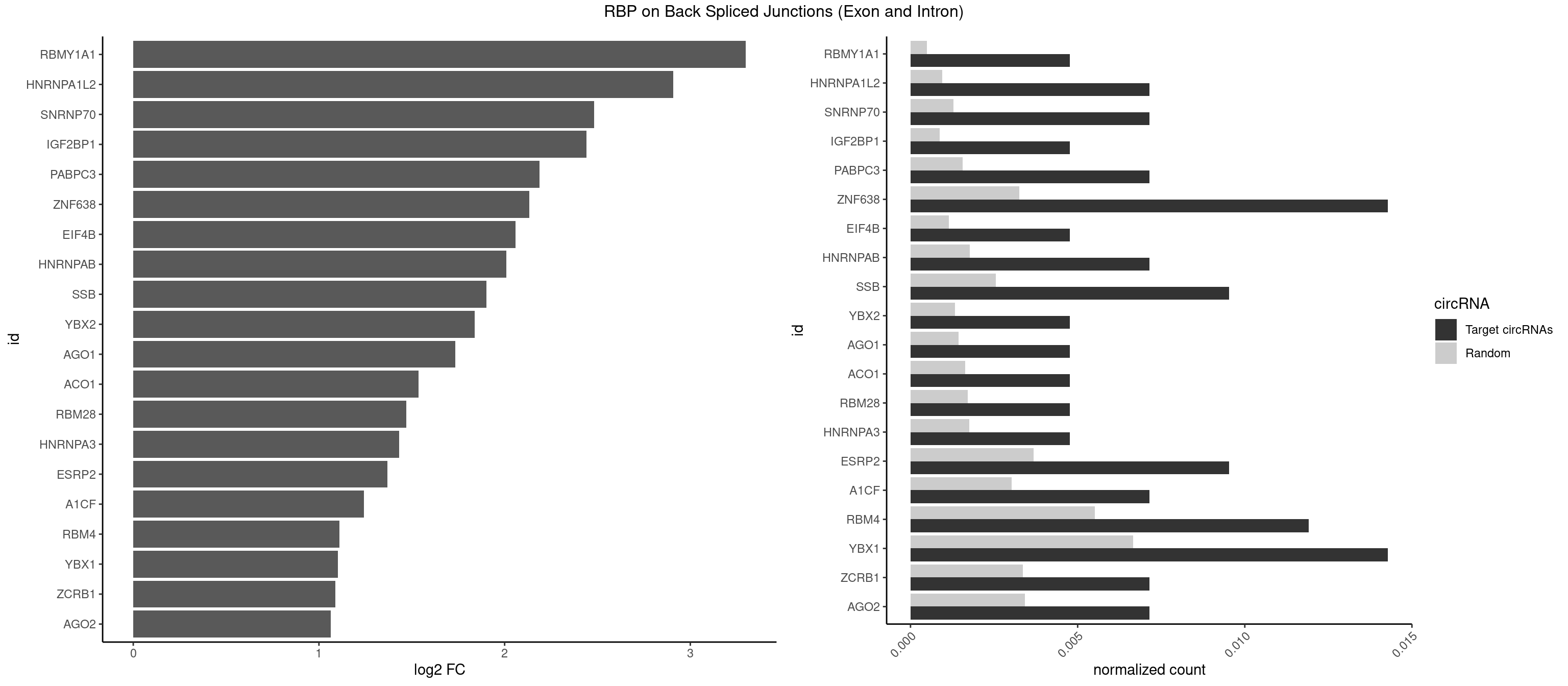
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | CCCGUU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZNF638 | 5 | 646 | 0.014285714 | 0.0032597743 | 2.131729 | GGUUCU,GUUCGU,GUUCUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | UCGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AAGACA,AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAACG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGGAU,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | UAAUUG,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUCUU,CCUUCC,CUUCCU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | CACACC,CAGCAA,CCAGCA,GCCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.