circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000197157:+:7:127887901:127929329
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000197157:+:7:127887901:127929329 | ENSG00000197157 | ENST00000354725 | + | 7 | 127887902 | 127929329 | 326 | AAACAUUGCUGAGGCUCUUGUCAGCAAAGGUCUAGCCACAGUGAUCAGAUACCGGCAGGAUGAUGACCAGAGAUCAUCACACUACGAUGAACUGCUUGCUGCAGAGGCCAGAGCUAUUAAGAAUGGCAAAGGAUUGCAUAGCAAGAAGGAAGUGCCUAUCCACCGUGUUGCAGAUAUAUCUGGGGAUACCCAAAAAGCAAAGCAGUUCCUGCCUUUUCUUCAGCGGGCAGGUCGUUCUGAAGCUGUGGUGGAAUACGUCUUCAGUGGUUCUCGUCUCAAACUCUAUUUGCCAAAGGAAACUUGCCUUAUCACCUUCUUGCUUGCAGAAACAUUGCUGAGGCUCUUGUCAGCAAAGGUCUAGCCACAGUGAUCAGAU | circ |
| ENSG00000197157:+:7:127887901:127929329 | ENSG00000197157 | ENST00000354725 | + | 7 | 127887902 | 127929329 | 22 | CUUGCUUGCAGAAACAUUGCUG | bsj |
| ENSG00000197157:+:7:127887901:127929329 | ENSG00000197157 | ENST00000354725 | + | 7 | 127887702 | 127887911 | 210 | AAAAAUGCUGAGCUAGGGAACUUUGUUAUUUAACCUUUUAUUUGGAUUUCUAGCUUAGCAACUCAUAGGUCUUGAAACCUUGGCUUCUCUCUCCCUCUGCCAGAAUGCGUUAAGACCUCCAGGUGCUAACUGUUAUUAUUUCUGUUGACUGAGCCCCAUAUGCACUUCUAGUGCUCACUGACCUAUCUUUUCUGUUGCAGAAACAUUGCU | ie_up |
| ENSG00000197157:+:7:127887901:127929329 | ENSG00000197157 | ENST00000354725 | + | 7 | 127929320 | 127929529 | 210 | UUGCUUGCAGGUAAGUCUUAUGUGUUACAUGUUACUCUGAGCUCAGAAGUCAUCCCUGGAAACCACAUACCCUCCUUUCCCCCUGUGUUCUAGAUAGGCCCUAGAGCCCACCAGCAUGCUCUUCCUCUCUGGUUGGGAUAUGCAAACACAGUUUCUAGAUUGGAUCCUCCUGUGCCUGCAGACCCAGCACUGCACCCCCUUUCUGCUCCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
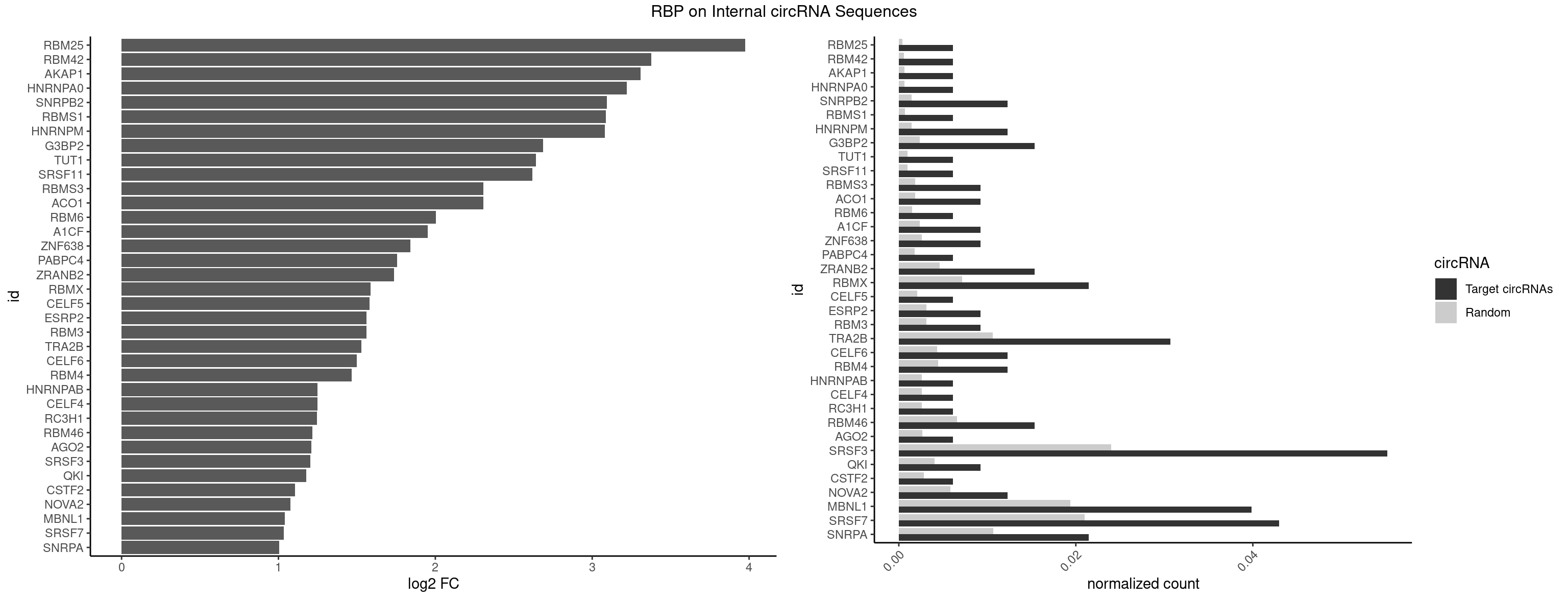
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 268 | 0.006134969 | 0.0003898359 | 3.976117 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| RBM42 | 1 | 407 | 0.006134969 | 0.0005912752 | 3.375154 | ACUACG | AACUAA,AACUAC,ACUAAG,ACUACG |
| AKAP1 | 1 | 426 | 0.006134969 | 0.0006188101 | 3.309488 | AUAUAU | AUAUAU,UAUAUA |
| HNRNPA0 | 1 | 453 | 0.006134969 | 0.0006579386 | 3.221031 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| SNRPB2 | 3 | 991 | 0.012269939 | 0.0014376103 | 3.093383 | AUUGCA,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBMS1 | 1 | 497 | 0.006134969 | 0.0007217036 | 3.087578 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| HNRNPM | 3 | 999 | 0.012269939 | 0.0014492040 | 3.081795 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 4 | 1644 | 0.015337423 | 0.0023839405 | 2.685636 | AGGAUG,AGGAUU,GGAUGA,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| TUT1 | 1 | 678 | 0.006134969 | 0.0009840095 | 2.640312 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SRSF11 | 1 | 688 | 0.006134969 | 0.0009985015 | 2.619220 | AAGAAG | AAGAAG |
| ACO1 | 2 | 1283 | 0.009202454 | 0.0018607779 | 2.306113 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS3 | 2 | 1283 | 0.009202454 | 0.0018607779 | 2.306113 | AUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBM6 | 1 | 1054 | 0.006134969 | 0.0015289102 | 2.004552 | UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| A1CF | 2 | 1642 | 0.009202454 | 0.0023810421 | 1.950426 | GAUCAG,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ZNF638 | 2 | 1773 | 0.009202454 | 0.0025708878 | 1.839752 | CGUUCU,GGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PABPC4 | 1 | 1251 | 0.006134969 | 0.0018144033 | 1.757561 | AAAAAG | AAAAAA,AAAAAG |
| ZRANB2 | 4 | 3173 | 0.015337423 | 0.0045997733 | 1.737421 | AAAGGU,AGGUCU,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| RBMX | 6 | 4925 | 0.021472393 | 0.0071387787 | 1.588734 | AAGAAG,AAGGAA,AGAAGG,AGGAAG,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| CELF5 | 1 | 1415 | 0.006134969 | 0.0020520728 | 1.579974 | GUGUUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ESRP2 | 2 | 2150 | 0.009202454 | 0.0031172377 | 1.561750 | GGGGAU,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM3 | 2 | 2152 | 0.009202454 | 0.0031201361 | 1.560410 | AAUACG,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| TRA2B | 9 | 7329 | 0.030674847 | 0.0106226650 | 1.529910 | AAGAAG,AAGAAU,AAGGAA,AGAAGG,AGGAAA,AGGAAG,GAAGGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| CELF6 | 3 | 2997 | 0.012269939 | 0.0043447134 | 1.497795 | GUGGUG,GUGUUG,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM4 | 3 | 3065 | 0.012269939 | 0.0044432593 | 1.465438 | CCUUCU,CUUCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| CELF4 | 1 | 1782 | 0.006134969 | 0.0025839306 | 1.247489 | GUGUUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPAB | 1 | 1782 | 0.006134969 | 0.0025839306 | 1.247489 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RC3H1 | 1 | 1786 | 0.006134969 | 0.0025897275 | 1.244256 | CCUUCU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBM46 | 4 | 4554 | 0.015337423 | 0.0066011240 | 1.216273 | AUGAUG,GAUCAU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| AGO2 | 1 | 1830 | 0.006134969 | 0.0026534924 | 1.209164 | AAGUGC | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SRSF3 | 17 | 16536 | 0.055214724 | 0.0239654858 | 1.204095 | ACUACG,AGAGAU,AUCAUC,CACUAC,CAGAGA,CAUCAC,CUACGA,CUCGUC,CUUCAG,CUUCUU,UACGAU,UCAUCA,UCGUUC,UCUUCA,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| QKI | 2 | 2805 | 0.009202454 | 0.0040664663 | 1.178243 | ACACUA,CACACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| CSTF2 | 1 | 1967 | 0.006134969 | 0.0028520334 | 1.105065 | GUGUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| NOVA2 | 3 | 4013 | 0.012269939 | 0.0058171047 | 1.076755 | AGAUCA,AUCACC,AUCAUC | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| MBNL1 | 12 | 13372 | 0.039877301 | 0.0193802045 | 1.040984 | CUGCCU,CUGCUU,CUUGCU,GCUUGC,UGCUGC,UUGCCA,UUGCCU,UUGCUG,UUGCUU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| SRSF7 | 13 | 14481 | 0.042944785 | 0.0209873716 | 1.032961 | AAAGGA,AAGAAG,ACUACG,AGAGAU,AGAUCA,AGGAAG,CAGAGA,CUACGA,GAGAUC,UACGAU,UCUUCA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| SNRPA | 6 | 7380 | 0.021472393 | 0.0106965744 | 1.005334 | AGCAGU,AUUGCA,GAUACC,UCCUGC,UUCCUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
RBP on BSJ (Exon Only)
Plot
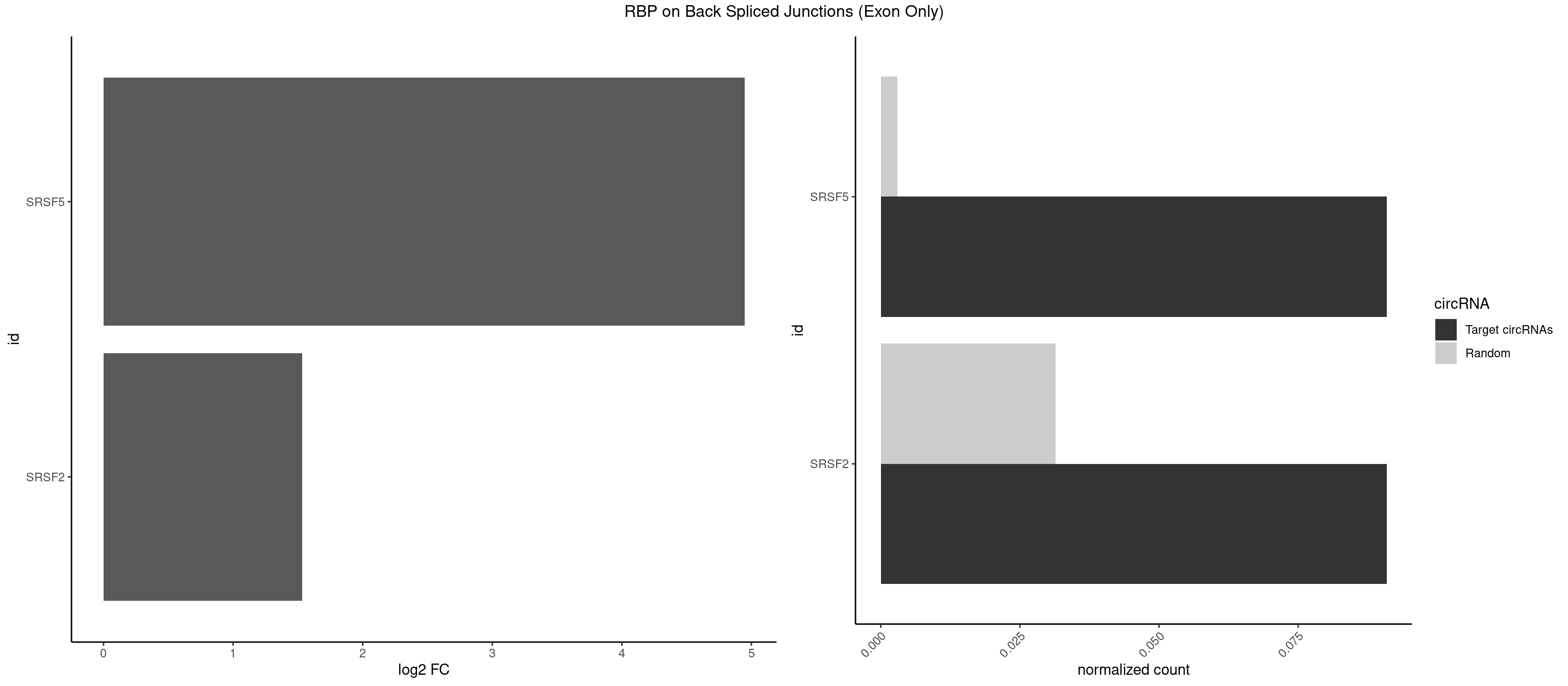
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF5 | 1 | 44 | 0.09090909 | 0.002947341 | 4.946939 | UGCAGA | AACAGC,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUC,CACGGA,CGCAGC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACGUA,UGCAGA,UGCAGC,UGCGGC |
| SRSF2 | 1 | 479 | 0.09090909 | 0.031438302 | 1.531901 | UGCAGA | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
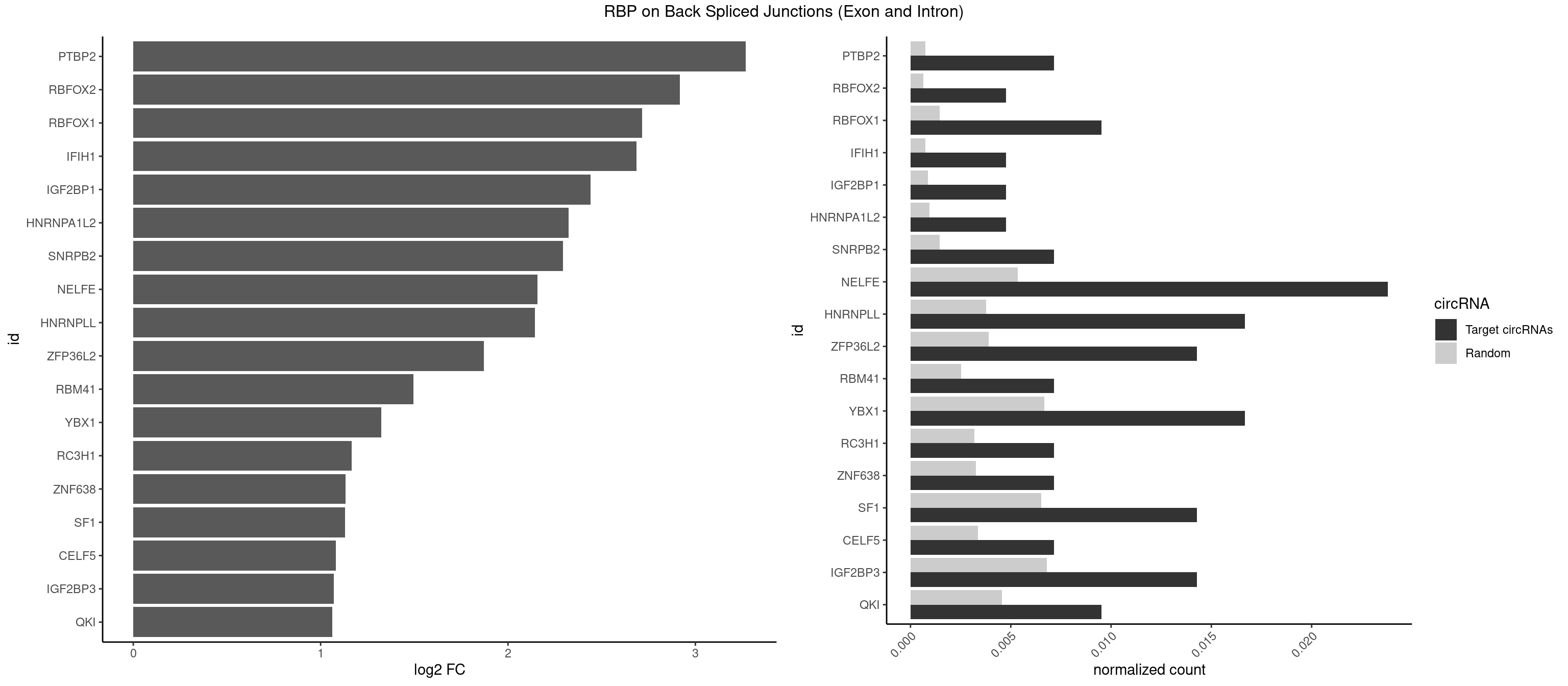
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 2 | 146 | 0.007142857 | 0.0007406288 | 3.269679 | CUCUCU | CUCUCU |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| RBFOX1 | 3 | 287 | 0.009523810 | 0.0014510278 | 2.714464 | AGCAUG,GCAUGC,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| NELFE | 9 | 1059 | 0.023809524 | 0.0053405885 | 2.156468 | CUCUCU,CUCUGG,CUGGUU,GCUAAC,UCUCUC,UCUCUG,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| HNRNPLL | 6 | 748 | 0.016666667 | 0.0037736800 | 2.142922 | ACAUAC,ACCACA,ACUGCA,CAAACA,CACUGC,GCAAAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| ZFP36L2 | 5 | 774 | 0.014285714 | 0.0039046755 | 1.871299 | UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACAUG,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| YBX1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | AACCAC,ACCACA,CCACCA,CCAGCA,GCCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUGG,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACUGAC,CACUGA,GCUAAC,UGCUAA,UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAACAC,AACACA,AACUCA,ACAUAC,CAAACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | ACUCAU,CUCAUA,UAACCU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.