circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000075151:-:1:21050865:21111383
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000075151:-:1:21050865:21111383 | ENSG00000075151 | MSTRG.488.20 | - | 1 | 21050866 | 21111383 | 266 | CAAUCCCACAGAGUAUUGAUGAGGAAACUGAAGUUUGGAGCGAUCACAUCAUUUUCCCAAGAAUUCCUAGAGGACCUGUGCAACAACCUCUUGAGGAUCGAAUCUUCACUCCCGCUGUCUCAGCAGUCUACAGCACGGUAACACAAGUGGCAAGACAGCCGGGAACCCCUACCCCAUCCCCUUAUUCAGCACAUGAAAUAAACAAGGGGCAUCCAAAUCUUGCGGCAACGCCCCCGGGACAUGCAUCGUCCCCUGGACUCUCUCAACAAUCCCACAGAGUAUUGAUGAGGAAACUGAAGUUUGGAGCGAUCACAUC | circ |
| ENSG00000075151:-:1:21050865:21111383 | ENSG00000075151 | MSTRG.488.20 | - | 1 | 21050866 | 21111383 | 22 | GACUCUCUCAACAAUCCCACAG | bsj |
| ENSG00000075151:-:1:21050865:21111383 | ENSG00000075151 | MSTRG.488.20 | - | 1 | 21111374 | 21111583 | 210 | AAGCUCUAAAGAAUGAAGUUAGGAGAUCCGUUGGUGCUCUUAAUUAGGAAUCUGGGAGAGAUGUCUUAAAGGCUGUGUCACCUGGAUAGUCCACGGAAAGGAAAACUCAUGAUCUUUAUGGAAGAGCUAUGUAUAAUAAUGAAAUACUGCUUAGUAUGUGCUAAGCCCAUCAUAAGCCUUAUUUUGCUAAUUCUUUCCAGCAAUCCCACA | ie_up |
| ENSG00000075151:-:1:21050865:21111383 | ENSG00000075151 | MSTRG.488.20 | - | 1 | 21050666 | 21050875 | 210 | ACUCUCUCAAGUAAGAUAAACCUUUUUUAAAAAAAUAAGAAAUGUCCCUGUAAAGGCAUCAUUUUUGUUUUUCUAAAGAGACUCCUAAGCAGUAACAUAAAACAUUACUUGUAUAGUAAUUUCAAGUUUGUCUUUGGCAUCUUUUCAGACUUAACCGUUCAUAAAGAUGGCUACUAACAAAUUUCACACAUUUCAUUUUGAAGUCAUUAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
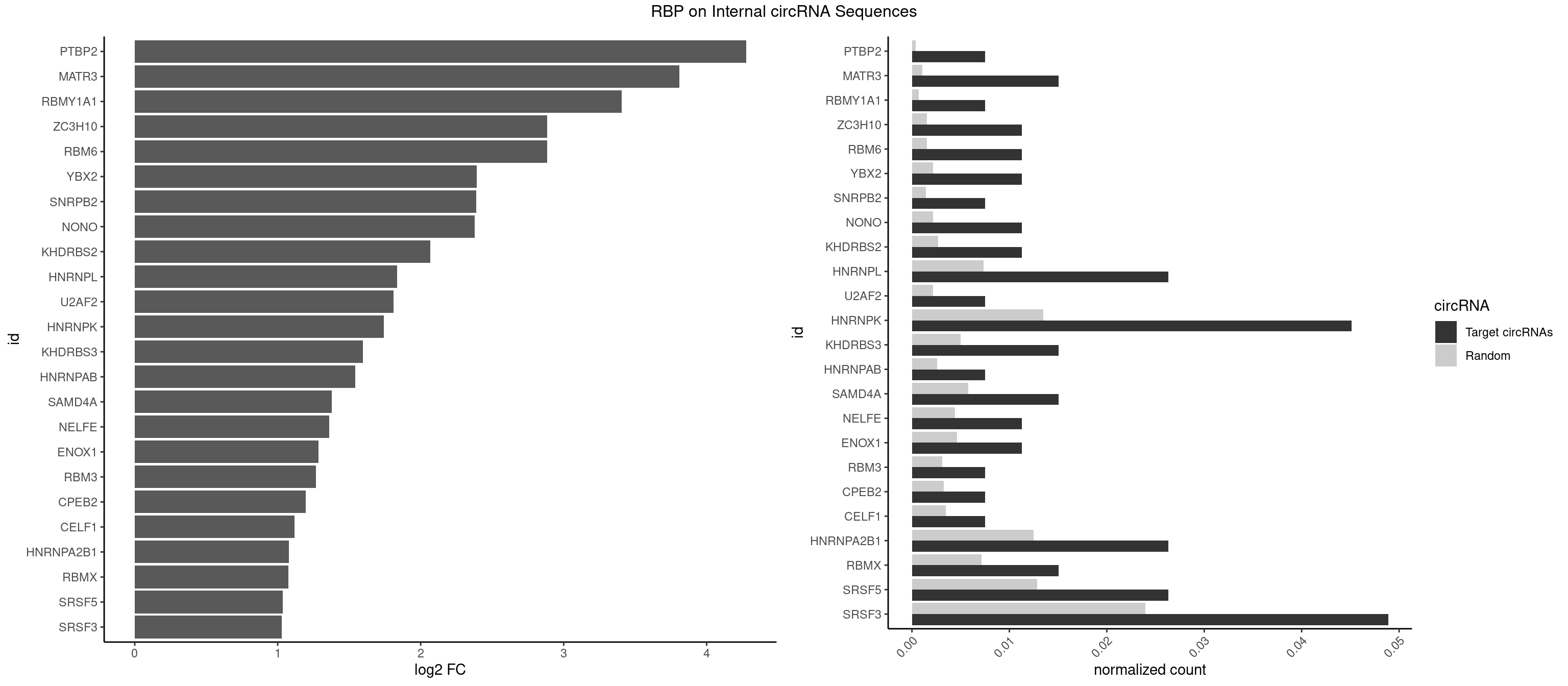
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 267 | 0.007518797 | 0.0003883867 | 4.274936 | CUCUCU | CUCUCU |
| MATR3 | 3 | 739 | 0.015037594 | 0.0010724109 | 3.809644 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMY1A1 | 1 | 489 | 0.007518797 | 0.0007101099 | 3.404388 | CAAGAC | ACAAGA,CAAGAC |
| ZC3H10 | 2 | 1053 | 0.011278195 | 0.0015274610 | 2.884329 | GAGCGA,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBM6 | 2 | 1054 | 0.011278195 | 0.0015289102 | 2.882961 | AUCCAA,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| YBX2 | 2 | 1480 | 0.011278195 | 0.0021462711 | 2.393632 | AACAAC,ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SNRPB2 | 1 | 991 | 0.007518797 | 0.0014376103 | 2.386829 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| NONO | 2 | 1498 | 0.011278195 | 0.0021723567 | 2.376203 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| KHDRBS2 | 2 | 1858 | 0.011278195 | 0.0026940701 | 2.065677 | AAUAAA,AUAAAC | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPL | 6 | 5085 | 0.026315789 | 0.0073706513 | 1.836065 | AAACAA,AAAUAA,AACACA,AAUAAA,ACACAA,CACAAG | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| U2AF2 | 1 | 1477 | 0.007518797 | 0.0021419234 | 1.811595 | UUUUCC | UUUUCC,UUUUUC,UUUUUU |
| HNRNPK | 11 | 9304 | 0.045112782 | 0.0134848428 | 1.742198 | ACCCCA,CAUCCC,CCAUCC,CCCCAU,CCCCUA,CCCCUU,GCCCCC,UCCCAA,UCCCAC,UCCCCU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| KHDRBS3 | 3 | 3429 | 0.015037594 | 0.0049707696 | 1.597033 | AAAUAA,AAUAAA,AUAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPAB | 1 | 1782 | 0.007518797 | 0.0025839306 | 1.540935 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SAMD4A | 3 | 3992 | 0.015037594 | 0.0057866714 | 1.377768 | CGGGAA,CGGGAC,CUGGAC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| NELFE | 2 | 3028 | 0.011278195 | 0.0043896388 | 1.361362 | CUCUCU,UCUCUC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| ENOX1 | 2 | 3195 | 0.011278195 | 0.0046316558 | 1.283936 | AAGACA,AGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RBM3 | 1 | 2152 | 0.007518797 | 0.0031201361 | 1.268893 | GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CPEB2 | 1 | 2261 | 0.007518797 | 0.0032780993 | 1.197642 | CAUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| CELF1 | 1 | 2391 | 0.007518797 | 0.0034664959 | 1.117024 | CUGUCU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPA2B1 | 6 | 8607 | 0.026315789 | 0.0124747476 | 1.076918 | AAGGGG,AGGGGC,CAAGAA,CAAGGG,CCAAGA,GGAACC | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| RBMX | 3 | 4925 | 0.015037594 | 0.0071387787 | 1.074825 | AUCCCA,AUCCCC,GUAACA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SRSF5 | 6 | 8869 | 0.026315789 | 0.0128544391 | 1.033662 | CACAGA,CACAUC,GAGGAA,UACAGC,UGCAUC,UGCGGC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| SRSF3 | 12 | 16536 | 0.048872180 | 0.0239654858 | 1.028055 | ACAUCA,AUCUUC,CACAUC,CAUCAU,CAUCGU,CCUCUU,CUACAG,CUUCAC,GAUCGA,UACAGC,UCGUCC,UCUUCA | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
RBP on BSJ (Exon Only)
Plot
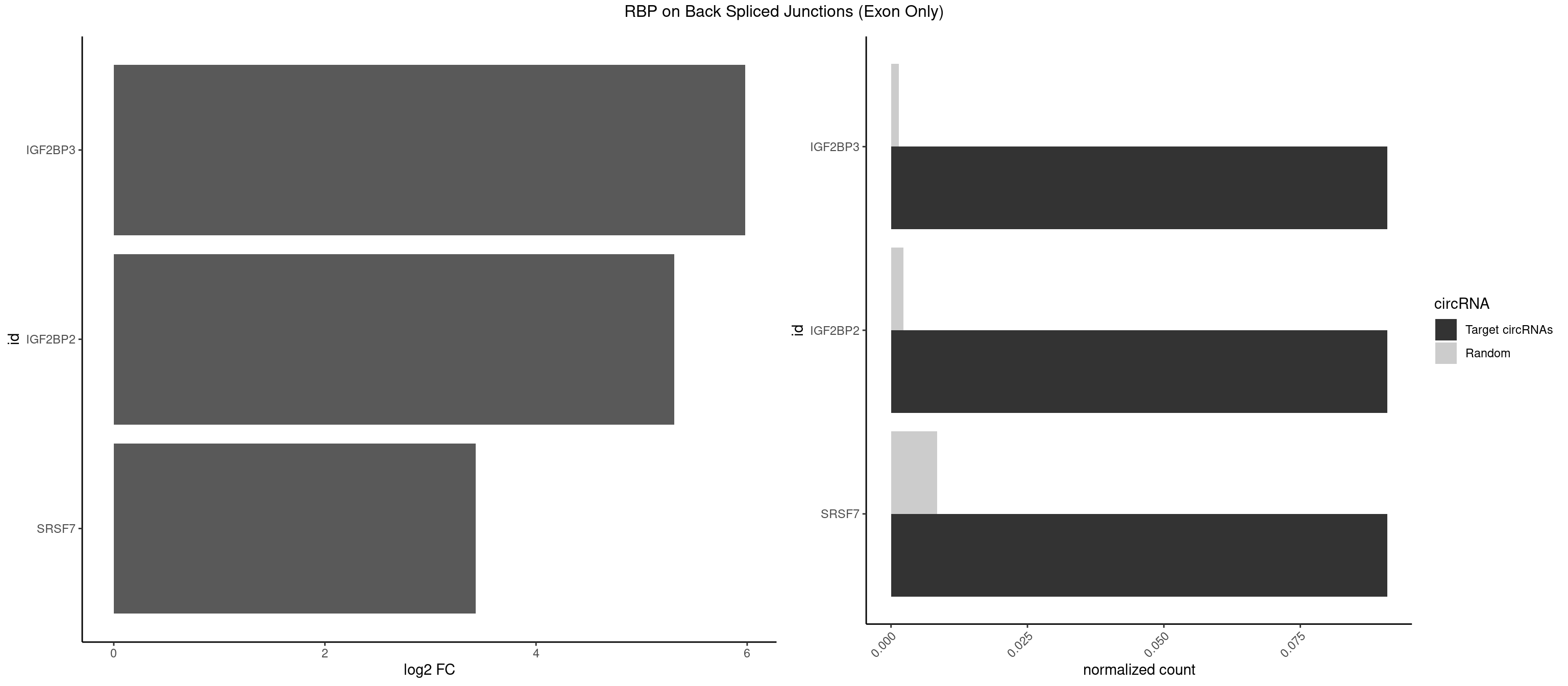
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGF2BP3 | 1 | 21 | 0.09090909 | 0.001440922 | 5.979360 | ACAAUC | AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,ACAAAC,ACAAUC,ACACUC,ACAUAC,CAAUCA,CACUCA,CAUACA,CAUUCA |
| IGF2BP2 | 1 | 34 | 0.09090909 | 0.002292376 | 5.309509 | ACAAUC | AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,ACAAAC,ACAAUC,ACACUC,ACAUAC,CAAAAC,CAAUAC,CAAUCA,CAAUUC,CACUCA,CAUACA,CAUUCA,CCAUAC,CCAUUC,GAAAAC,GAACAC,GAAUAC,GAAUUC |
| SRSF7 | 1 | 128 | 0.09090909 | 0.008449044 | 3.427565 | UCAACA | AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACUACG,AGAAGA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CGAAUG,CUCUUC,CUGAGA,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UAGAGA,UCUUCA,UGAGAG,UGGACA |
RBP on BSJ (Exon and Intron)
Plot
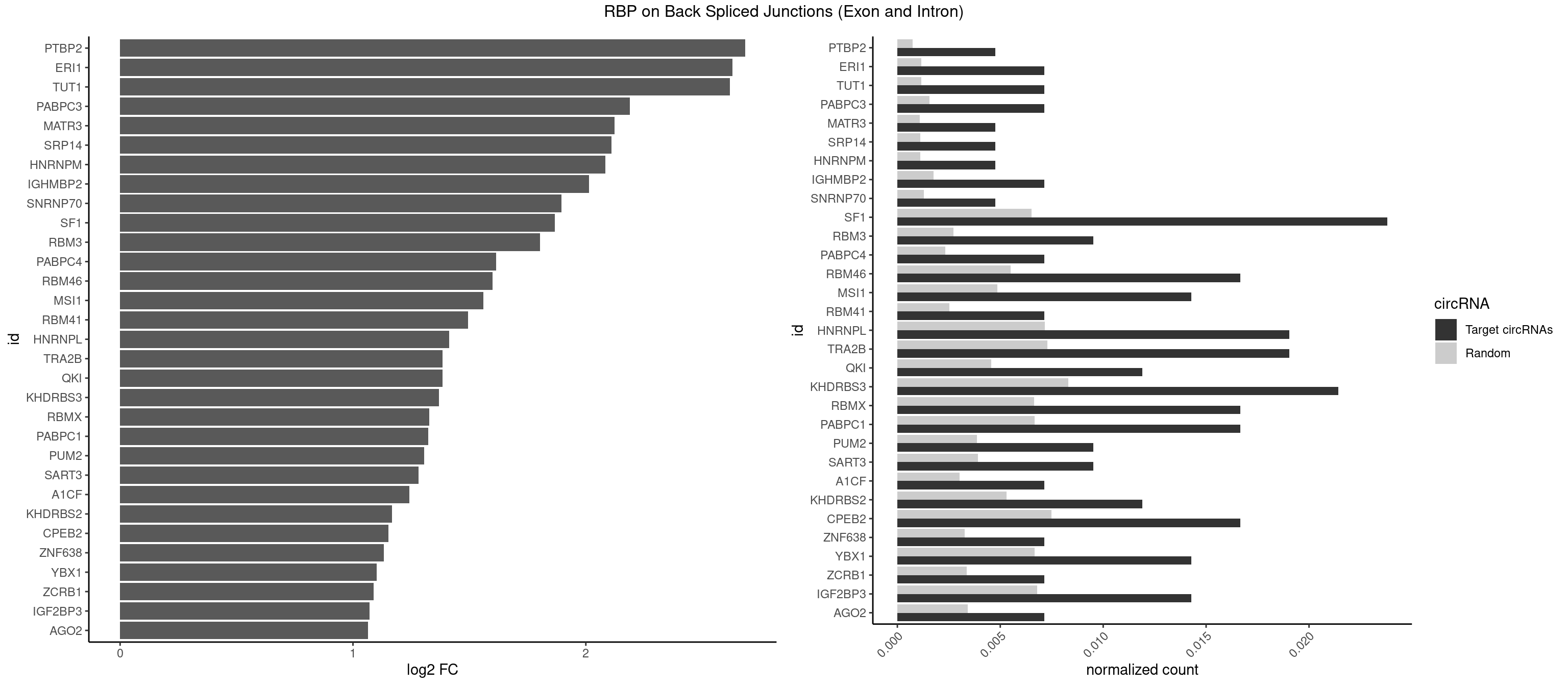
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CCUGUA | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| IGHMBP2 | 2 | 350 | 0.007142857 | 0.0017684401 | 2.014024 | AAAAAA | AAAAAA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| SF1 | 9 | 1294 | 0.023809524 | 0.0065245869 | 1.867580 | ACUAAC,AGUAAC,AGUAAG,CUAACA,UAACAA,UACUAA,UAGUAA,UGCUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AAUACU,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA | AAAAAA,AAAAAG |
| RBM46 | 6 | 1091 | 0.016666667 | 0.0055018138 | 1.598986 | AAUGAA,AUCAUA,AUCAUU,AUGAAA,AUGAAG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| MSI1 | 5 | 961 | 0.014285714 | 0.0048468360 | 1.559458 | AGUAAG,AGUUAG,UAGGAA,UAGGAG,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPL | 7 | 1419 | 0.019047619 | 0.0071543732 | 1.412713 | AAAUAA,AAAUAC,AACAAA,ACAUAA,CACACA,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| TRA2B | 7 | 1447 | 0.019047619 | 0.0072954454 | 1.384543 | AAAGAA,AAGAAU,AAGGAA,AAUAAG,AGGAAA,AUAAGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACUAAC,ACUCAU,AUCAUA,CUAACA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| KHDRBS3 | 8 | 1646 | 0.021428571 | 0.0082980653 | 1.368689 | AAAUAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,GAUAAA,UAAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| RBMX | 6 | 1316 | 0.016666667 | 0.0066354293 | 1.328704 | AAGGAA,AAGUAA,AGUAAC,AUCCCA,GUAACA,UAACAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| PABPC1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | AAAAAA,ACAAAU,ACUAAC,CUAACA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UGUAAA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | AUAAAA,AUAAAC,GAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| CPEB2 | 6 | 1487 | 0.016666667 | 0.0074969770 | 1.152585 | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | CGUUGG,GUUGGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | CAGCAA,CAUCAU,CCAGCA,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | ACUUAA,GACUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAACA,AAACUC,AAAUAC,AACUCA,CACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.