circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000044115:+:5:138827514:138887642
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000044115:+:5:138827514:138887642 | ENSG00000044115 | ENST00000627109 | + | 5 | 138827515 | 138887642 | 438 | AAACAAAUCAUUGUGGACCCCUUGAGCUUCAGCGAGGAGCGCUUUAGGCCUUCCCUGGAGGAGCGUCUGGAAAGCAUCAUUAGUGGGGCUGCCUUGAUGGCCGACUCGUCCUGCACGCGUGAUGACCGUCGUGAGCGAAUUGUGGCAGAGUGUAAUGCUGUCCGCCAGGCCCUGCAGGACCUGCUUUCGGAGUACAUGGGCAAUGCUGGACGUAAAGAAAGAAGUGAUGCACUCAAUUCUGCAAUAGAUAAAAUGACCAAGAAGACCAGGGACUUGCGUAGACAGCUCCGCAAAGCUGUCAUGGACCACGUUUCAGAUUCUUUCCUGGAAACCAAUGUUCCACUUUUGGUAUUGAUUGAAGCUGCAAAGAAUGGAAAUGAGAAAGAAGUUAAGGAGUAUGCCCAAGUUUUCCGUGAACAUGCCAACAAAUUGAUUGAGAAACAAAUCAUUGUGGACCCCUUGAGCUUCAGCGAGGAGCGCUUUAGGCC | circ |
| ENSG00000044115:+:5:138827514:138887642 | ENSG00000044115 | ENST00000627109 | + | 5 | 138827515 | 138887642 | 22 | AAUUGAUUGAGAAACAAAUCAU | bsj |
| ENSG00000044115:+:5:138827514:138887642 | ENSG00000044115 | ENST00000627109 | + | 5 | 138827315 | 138827524 | 210 | CUUCCACUGCUUUCUUUGAGGAGUGAUCAACAGUAGGCCAUCUUCUGUGGGACAGCCCCUUACCUGCUAAGAAGUAUAACUUUCUUUCAGAUGUUGGAAUUUUUGUGUUAAAUCUUGAUAUUACUAAUAACACUUUUCUCACCUUGAAUAAAGAAGGGAACAGAGAUGAGUACUAACAUUCGGUAAUACUUUCUCUGCAGAAACAAAUCA | ie_up |
| ENSG00000044115:+:5:138827514:138887642 | ENSG00000044115 | ENST00000627109 | + | 5 | 138887633 | 138887842 | 210 | AUUGAUUGAGGUAAGUGAAUUAGCAGUUUCAUUGACUUGUAGGCAACUUGGUGAAGUGUGGUGAGUUUUAAUCACAUUAUUUAAUCAGUUAAAAUUUGUAAGGGCUCGCUUAACAUACAACAUGUCUGAGAUGAUAGUCUUUCAUUAUCACUUAACAUCUAAGGAAAACUACCAUUUUGGUCACUUUACAUUAACUAUGUACGUUAUUAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
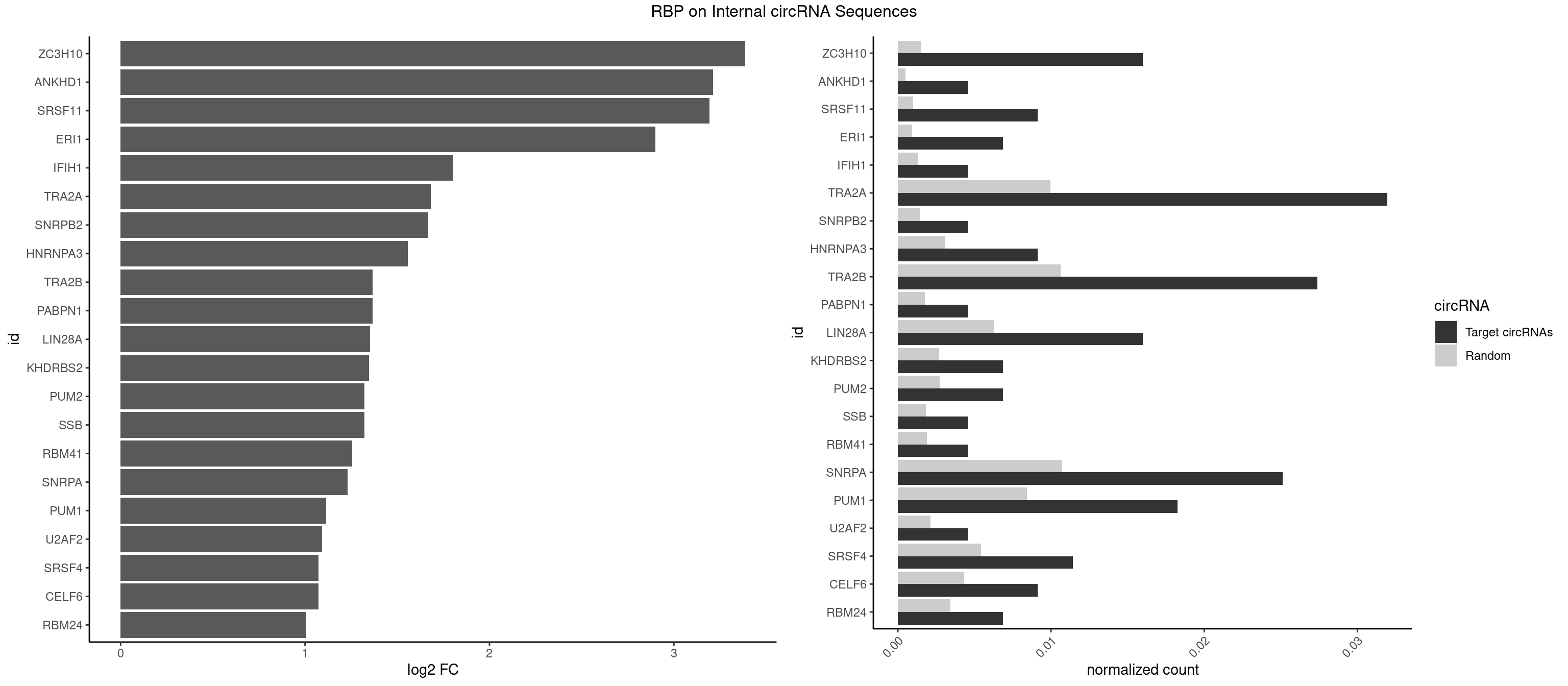
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZC3H10 | 6 | 1053 | 0.015981735 | 0.0015274610 | 3.387217 | CAGCGA,GAGCGA,GAGCGC,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| ANKHD1 | 1 | 339 | 0.004566210 | 0.0004927293 | 3.212130 | GACGUA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SRSF11 | 3 | 688 | 0.009132420 | 0.0009985015 | 3.193161 | AAGAAG | AAGAAG |
| ERI1 | 2 | 632 | 0.006849315 | 0.0009173461 | 2.900422 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| IFIH1 | 1 | 904 | 0.004566210 | 0.0013115296 | 1.799747 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| TRA2A | 13 | 6871 | 0.031963470 | 0.0099589296 | 1.682361 | AAAGAA,AAGAAA,AAGAAG,AGAAAG,AGAAGA,GAAAGA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| SNRPB2 | 1 | 991 | 0.004566210 | 0.0014376103 | 1.667325 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPA3 | 3 | 2140 | 0.009132420 | 0.0031027457 | 1.557452 | AAGGAG,AGGAGC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| TRA2B | 11 | 7329 | 0.027397260 | 0.0106226650 | 1.366886 | AAAGAA,AAGAAG,AAGAAU,AGAAGA,GAAAGA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| PABPN1 | 1 | 1222 | 0.004566210 | 0.0017723764 | 1.365312 | AGAAGA | AAAAGA,AGAAGA |
| LIN28A | 6 | 4315 | 0.015981735 | 0.0062547643 | 1.353397 | AGGAGU,CGGAGU,GGAGGA,GGAGUA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| KHDRBS2 | 2 | 1858 | 0.006849315 | 0.0026940701 | 1.346172 | AUAAAA,GAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| PUM2 | 2 | 1890 | 0.006849315 | 0.0027404447 | 1.321550 | GUACAU,UAGAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SSB | 1 | 1260 | 0.004566210 | 0.0018274462 | 1.321168 | UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM41 | 1 | 1318 | 0.004566210 | 0.0019115000 | 1.256292 | UACAUG | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SNRPA | 10 | 7380 | 0.025114155 | 0.0106965744 | 1.231352 | ACCUGC,AUGCAC,AUGCUG,CCUGCU,GUAUGC,UCCUGC,UGCACG,UUCCUG,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| PUM1 | 7 | 5823 | 0.018264840 | 0.0084401638 | 1.113726 | AAUGUU,AAUUGU,AGAUAA,GAAUUG,GUACAU,UAGAUA,UGUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| U2AF2 | 1 | 1477 | 0.004566210 | 0.0021419234 | 1.092090 | UUUUCC | UUUUCC,UUUUUC,UUUUUU |
| SRSF4 | 4 | 3740 | 0.011415525 | 0.0054214720 | 1.074241 | AAGAAG,AGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| CELF6 | 3 | 2997 | 0.009132420 | 0.0043447134 | 1.071736 | GUGAUG,GUGGGG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM24 | 2 | 2357 | 0.006849315 | 0.0034172229 | 1.003135 | AGAGUG,GAGUGU | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
RBP on BSJ (Exon Only)
Plot
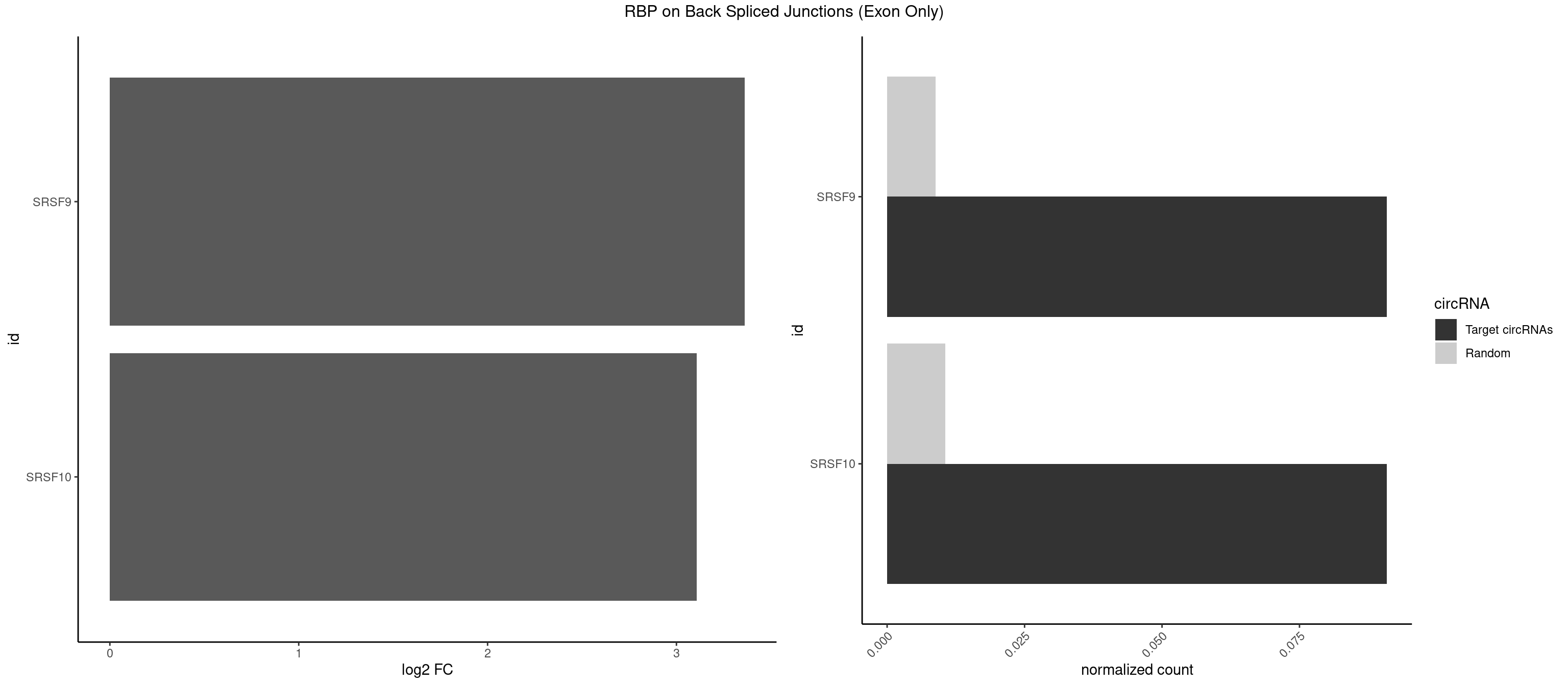
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF9 | 1 | 134 | 0.09090909 | 0.008842023 | 3.361976 | UGAGAA | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
| SRSF10 | 1 | 160 | 0.09090909 | 0.010544931 | 3.107875 | GAGAAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon and Intron)
Plot

Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF11 | 2 | 82 | 0.007142857 | 0.0004181782 | 4.094312 | AAGAAG | AAGAAG |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| MATR3 | 3 | 216 | 0.009523810 | 0.0010933091 | 3.122837 | AAUCUU,AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| YBX2 | 3 | 263 | 0.009523810 | 0.0013301088 | 2.839994 | AACAUC,ACAACA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | GUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | GAGGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | AUACUU,UACAUU,UACUUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| QKI | 7 | 904 | 0.019047619 | 0.0045596534 | 2.062615 | ACUAAC,ACUAAU,AUCUAA,AUUAAC,CUAACA,UAAUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGG,UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | ACUUAA,AUUUAA,GAAUUA,GCUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AAACUA,AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | AGUAUA,AUCAGU,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| TRA2B | 8 | 1447 | 0.021428571 | 0.0072954454 | 1.554468 | AAAGAA,AAGAAG,AAGGAA,AGAAGG,AGGAAA,GAAUUA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SF1 | 7 | 1294 | 0.019047619 | 0.0065245869 | 1.545652 | ACUAAC,ACUAAU,AUUAAC,CUAACA,UACUAA,UGCUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| PABPC1 | 7 | 1321 | 0.019047619 | 0.0066606207 | 1.515882 | ACAAAU,ACUAAC,ACUAAU,CAAAUC,CUAACA,CUAAUA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| SNRPA | 10 | 1947 | 0.026190476 | 0.0098145909 | 1.416042 | ACCUGC,AGCAGU,AGUAGG,CAGUAG,CCUGCU,CUGCUA,GUAGGC,UACCUG,UUACCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AAGAAG | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPLL | 3 | 748 | 0.009523810 | 0.0037736800 | 1.335567 | ACAUAC,CACUGC,CAUACA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| CELF6 | 5 | 1196 | 0.014285714 | 0.0060308343 | 1.244144 | GUGGUG,GUGUGG,UGUGGG,UGUGGU,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBMX | 5 | 1316 | 0.014285714 | 0.0066354293 | 1.106311 | AAGAAG,AAGGAA,AAGUGU,AGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | AGUGUG,GAGUGA,GUGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUGG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.