circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000033867:-:3:27383152:27452498
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000033867:-:3:27383152:27452498 | ENSG00000033867 | ENST00000454389 | - | 3 | 27383153 | 27452498 | 3530 | GGUCCUGAUGAAGAAGCUGUUGUGGAUCUUGGCAAAACUAGCUCAACUGUGAACACCAAGUUUGAAAAAGAAGAACUAGAAAGUCAUAGAGCUGUAUAUAUUGGUGUUCACGUCCCGUUUAGUAAAGAGAGUCGUCGGCGUCAUAGGCAUCGCGGACACAAACAUCACCACCGGAGAAGAAAAGAUAAAGAAUCAGAUAAAGAAGAUGGACGGGAAUCUCCUUCUUAUGAUACACCAUCCCAGAGAGUUCAGUUUAUCCUUGGUACUGAAGAUGAUGAUGAAGAACAUAUUCCCCAUGAUCUCUUCACGGAAAUGGAUGAACUGUGUUACAGAGAUGGAGAAGAAUAUGAAUGGAAAGAAACUGCUAGAUGGCUGAAAUUUGAAGAGGAUGUUGAAGAUGGCGGUGACCGAUGGAGUAAACCUUAUGUGGCAACUCUCUCUUUGCACAGUCUUUUUGAACUAAGGAGUUGCAUCCUCAAUGGAACAGUCAUGCUGGAUAUGAGAGCAAGCACUCUAGAUGAAAUAGCAGAUAUGGUAUUAGACAACAUGAUAGCUUCUGGCCAAUUAGACGAGUCCAUACGAGAGAAUGUCAGAGAAGCUCUUCUGAAGAGACAUCAUCAUCAGAAUGAGAAAAGAUUCACCAGUCGGAUUCCUCUUGUUCGAUCUUUUGCAGAUAUAGGCAAGAAACAUUCUGACCCUCACUUGCUUGAAAGGAAUGGGGAAGGCCUUUCAGCCUCCCGCCACUCUUUGCGAACAGGUCUGUCUGCCUCAAACCUUUCCUUGAGAGGAGAAUCACCUUUAUCUCUUCUUCUUGGUCAUCUUCUUCCUUCUUCAAGAGCUGGAACCCCUGCAGGCUCAAGGUGUACAACCCCAGUACCCACCCCUCAAAACAGUCCUCCUUCUAGCCCUAGCAUCAGCCGCCUGACCUCCAGAAGUUCCCAAGAGAGUCAGCGUCAGGCCCCAGAACUACUGGUUUCACCUGCCAGUGAUGAUAUUCCCACAGUAGUAAUUCAUCCGCCUGAGGAAGACUUAGAAGCAGCGCUGAAAGGCGAGGAGCAGAAGAAUGAGGAAAAUGUUGACUUAACUCCAGGUAUUUUGGCCUCUCCCCAGUCUGCUCCUGGAAACUUGGACAAUAGUAAAAGUGGAGAAAUUAAAGGUAAUGGAAGUGGUGGAAGCAGAGAAAAUAGUACUGUUGACUUCAGCAAGGUUGAUAUGAAUUUCAUGAGAAAAAUUCCUACGGGUGCUGAGGCAUCCAACGUCCUGGUGGGCGAAGUAGACUUUUUGGAAAGGCCAAUAAUUGCAUUUGUGAGACUGGCUCCUGCUGUCCUCCUUACAGGGUUGACUGAGGUCCCUGUUCCAACCAGGUUUUUGUUUUUGUUAUUGGGUCCAGCGGGCAAGGCACCACAGUACCAUGAAAUUGGACGAUCAAUAGCCACUCUCAUGACAGAUGAGAUUUUCCAUGAUGUAGCUUAUAAAGCAAAAGACAGAAAUGACCUCUUAUCUGGAAUUGAUGAAUUUUUAGAUCAAGUAACUGUCCUACCUCCAGGAGAGUGGGAUCCUUCUAUACGCAUAGAACCACCAAAAAGUGUCCCUUCUCAGGAAAAGAGAAAGAUUCCUGUGUUUCACAAUGGAUCUACCCCCACACUGGGUGAGACUCCUAAAGAGGCCGCUCAUCAUGCUGGGCCUGAGCUACAGAGGACUGGACGGCUUUUUGGUGGUUUGAUACUUGACAUCAAAAGGAAAGCACCUUUUUUCUUGAGUGACUUCAAGGAUGCAUUAAGCCUGCAGUGCCUGGCCUCGAUUCUUUUCCUAUACUGUGCCUGUAUGUCUCCUGUAAUCACUUUUGGAGGGCUGCUUGGAGAAGCUACAGAAGGCAGAAUAAGUGCAAUAGAGUCUCUUUUUGGAGCAUCAUUAACUGGGAUUGCCUAUUCAUUGUUUGCUGGGCAACCUCUAACAAUAUUGGGGAGCACAGGUCCAGUUCUAGUGUUUGAAAAAAUUUUAUAUAAAUUCUGCAGAGAUUAUCAACUUUCUUAUCUGUCUUUAAGAACCAGUAUUGGUCUGUGGACUUCUUUUUUGUGCAUUGUUUUGGUUGCAACAGAUGCAAGCAGCCUUGUGUGUUAUAUUACUCGAUUUACAGAAGAGGCUUUUGCAGCCCUUAUUUGCAUCAUAUUCAUCUACGAGGCUUUGGAGAAGCUCUUUGAUUUAGGAGAAACAUAUGCAUUUAAUAUGCACAACAACUUAGAUAAACUGACCAGCUACUCAUGUGUAUGUACUGAACCUCCAAACCCCAGCAAUGAAACUCUAGCACAAUGGAAGAAAGAUAAUAUAACAGCACACAAUAUUUCCUGGAGAAAUCUUACUGUUUCUGAAUGUAAAAAACUUCGUGGUGUAUUCUUGGGGUCAGCUUGUGGUCAUCAUGGACCUUAUAUUCCAGAUGUGCUCUUUUGGUGUGUCAUCUUGUUUUUCACAACAUUUUUUCUGUCUUCAUUCCUCAAGCAAUUUAAGACCAAGCGUUACUUUCCUACCAAGGUGCGAUCGACAAUCAGUGAUUUUGCUGUAUUUCUCACAAUAGUAAUAAUGGUUACAAUUGACUACCUUGUAGGAGUUCCAUCUCCUAAACUUCAUGUUCCUGAAAAAUUUGAGCCUACUCAUCCAGAGAGAGGGUGGAUCAUAAGCCCACUGGGAGAUAAUCCUUGGUGGACCUUAUUAAUAGCUGCUAUUCCUGCUUUGCUUUGUACCAUUCUCAUCUUUAUGGAUCAACAAAUCACAGCUGUAAUUAUAAACAGAAAGGAACACAAAUUGAAGAAAGGAGCUGGCUAUCACCUUGAUUUGCUCAUGGUUGGCGUUAUGUUGGGAGUUUGCUCUGUCAUGGGACUUCCAUGGUUUGUGGCUGCAACAGUGUUGUCAAUAAGUCAUGUCAACAGCUUAAAAGUUGAAUCUGAAUGUUCUGCUCCAGGGGAACAACCCAAGUUUUUGGGAAUUCGUGAACAGCGGGUUACAGGGCUAAUGAUUUUUAUUCUAAUGGGCCUCUCUGUGUUCAUGACUUCAGUCCUAAAGUUUAUUCCAAUGCCUGUUCUGUAUGGUGUUUUCCUUUAUAUGGGAGUUUCCUCAUUAAAAGGAAUCCAGUUAUUUGACCGUAUAAAAUUAUUUGGAAUGCCUGCUAAGCAUCAGCCUGAUUUGAUAUACCUCCGUUAUGUGCCGCUCUGGAAGGUCCAUAUUUUCACAGUCAUUCAGCUUACUUGUUUGGUCCUUUUAUGGGUGAUAAAAGUUUCAGCUGCUGCAGUGGUUUUUCCCAUGAUGGUUCUUGCAUUAGUGUUUGUGCGCAAACUCAUGGACCUGUGUUUCACGAAGAGAGAACUUAGUUGGCUUGAUGAUCUUAUGCCAGAAAGUAAGAAAAAGAAAGAAGAUGACAAAAAGAAAAAAGAGAAAGAGGAAGCUGAACGGAUGCUUCAAGAUGAUGAUGAUACUGUGCACCUUCCAUUUGAAGGGGGAAGUCUCUUGCAAAUUCCAGUCAAGGCCCUAAAAUAUAGGGUCCUGAUGAAGAAGCUGUUGUGGAUCUUGGCAAAACUAGCUCAACUGU | circ |
| ENSG00000033867:-:3:27383152:27452498 | ENSG00000033867 | ENST00000454389 | - | 3 | 27383153 | 27452498 | 22 | CUAAAAUAUAGGGUCCUGAUGA | bsj |
| ENSG00000033867:-:3:27383152:27452498 | ENSG00000033867 | ENST00000454389 | - | 3 | 27452489 | 27452698 | 210 | CUCUGGGAUUUUUGAAUGGUCUUCUGUGGAUUUAAAAAGCUUGUAAACAUAAUUUUAAAAGAAUGCAAUUAGCUGUAAUGAGUUCAGUUAUUUGAGAGAAAGCAGUCUGUUUUAUAUUUUGUUGCCAUUUCAAAGGAUGCAUAUAAUAGGUCCUCAAUAGAUUGUGAUCUCAUGAGUCAAUGAGAAUAAUUUGUUUUUAGGGUCCUGAUG | ie_up |
| ENSG00000033867:-:3:27383152:27452498 | ENSG00000033867 | ENST00000454389 | - | 3 | 27382953 | 27383162 | 210 | UAAAAUAUAGGUGAGAUAUGACUUUAUGCUCUAAAACUUUUAUAUGCAUGUCAAAUGUAUUAAUAAGUCAUAUAAUGCUUCUUUAAAGUGAUGAUUUCUAUUUUAUAAAAGAUGAUACAAUCUGCAUAUGGUGUAUCAACUAAAGGAGGGCUUCACACAAUUUGAAAAGGAAGUCCCUUGCUCCUUUCCCACCCAAGCUGGACUUGGACU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
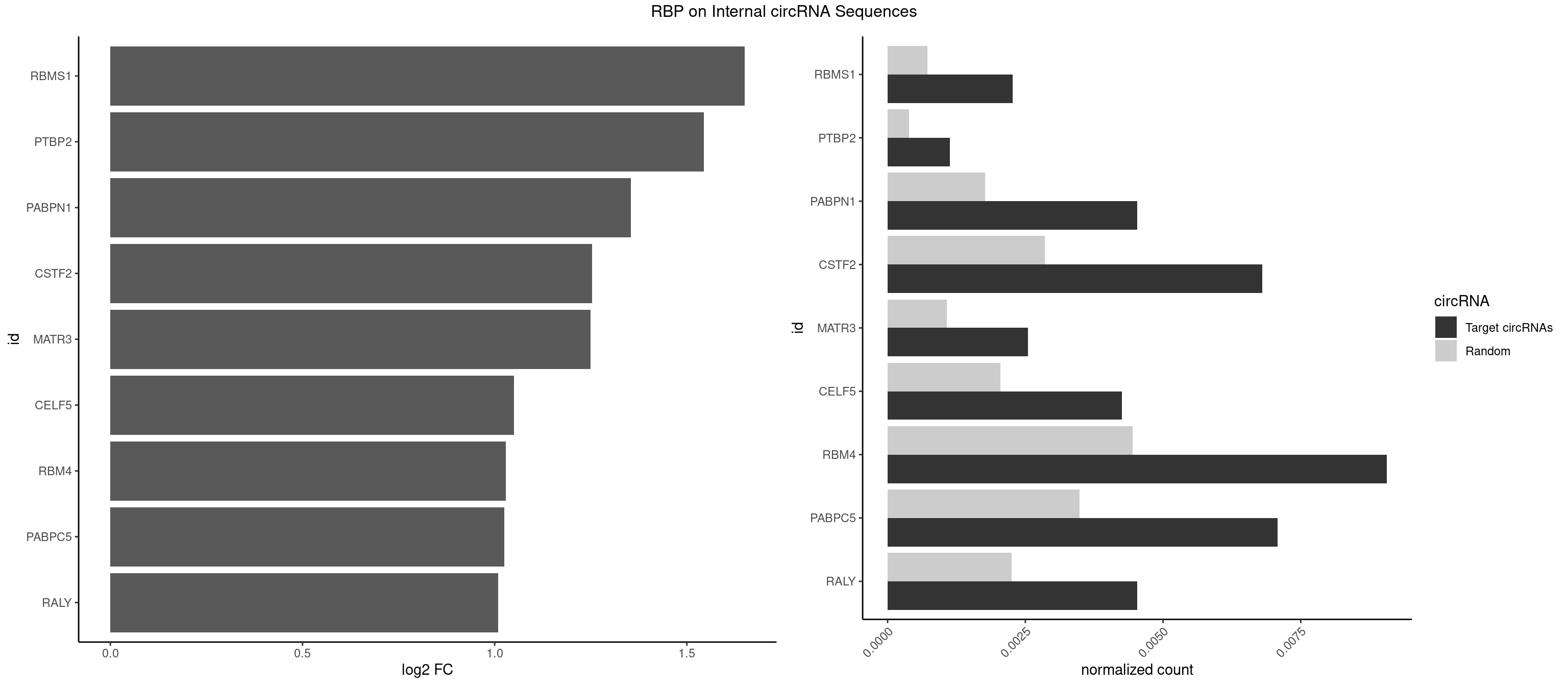
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMS1 | 7 | 497 | 0.002266289 | 0.0007217036 | 1.650854 | AUAUAC,AUAUAG,GAUAUA,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| PTBP2 | 3 | 267 | 0.001133144 | 0.0003883867 | 1.544766 | CUCUCU | CUCUCU |
| PABPN1 | 15 | 1222 | 0.004532578 | 0.0017723764 | 1.354647 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| CSTF2 | 23 | 1967 | 0.006798867 | 0.0028520334 | 1.253303 | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| MATR3 | 8 | 739 | 0.002549575 | 0.0010724109 | 1.249399 | AAUCUU,AUCUUA,AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| CELF5 | 14 | 1415 | 0.004249292 | 0.0020520728 | 1.050141 | GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM4 | 31 | 3065 | 0.009065156 | 0.0044432593 | 1.028713 | CCUCUU,CCUUCC,CCUUCU,CUCUUU,CUUCCU,CUUCUU,UCCUUC,UUCCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| PABPC5 | 24 | 2400 | 0.007082153 | 0.0034795387 | 1.025292 | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RALY | 15 | 1553 | 0.004532578 | 0.0022520629 | 1.009085 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
RBP on BSJ (Exon Only)
Plot
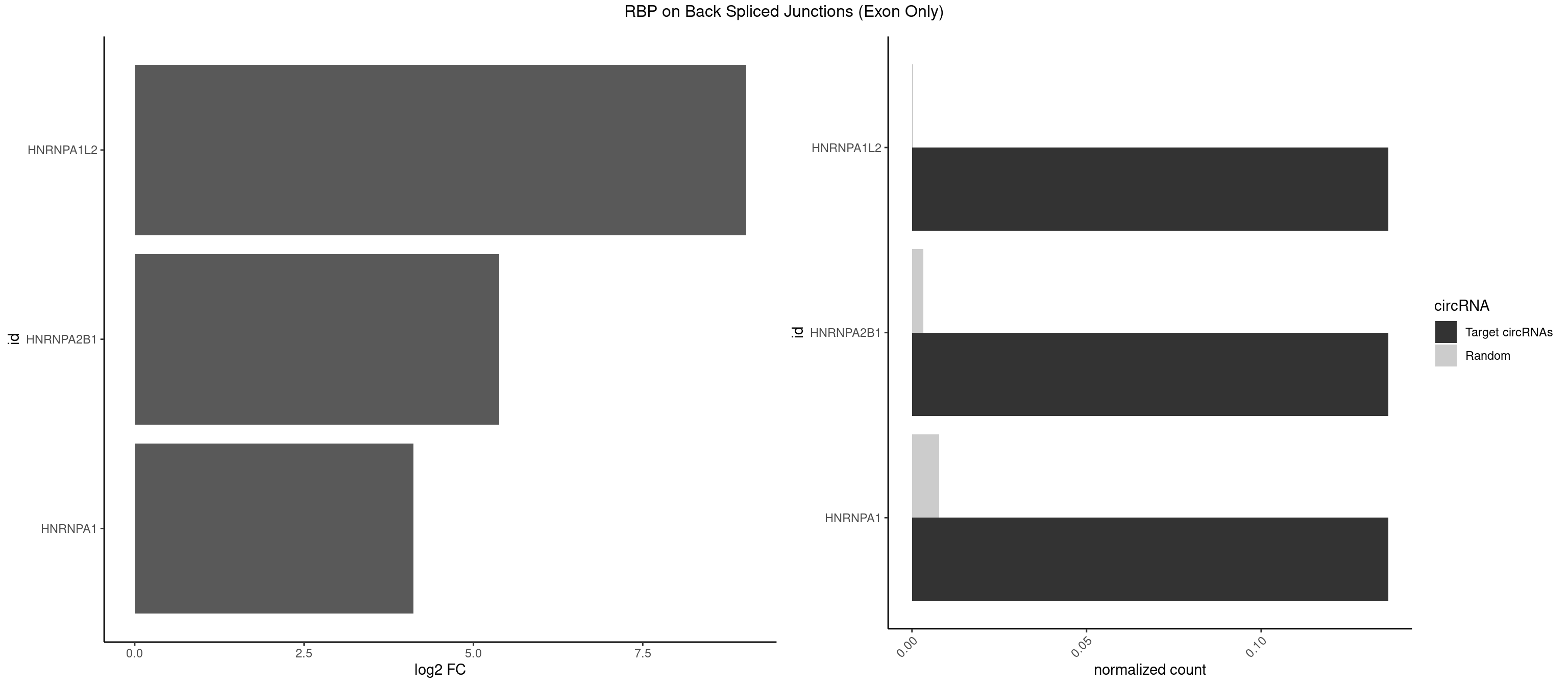
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 2 | 3 | 0.1363636 | 0.0002619859 | 9.023754 | AUAGGG,UAGGGU | AUAGGG,UUAGGG |
| HNRNPA2B1 | 2 | 49 | 0.1363636 | 0.0032748232 | 5.379898 | AUAGGG,UAGGGU | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| HNRNPA1 | 2 | 119 | 0.1363636 | 0.0078595756 | 4.116864 | AUAGGG,UAGGGU | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
RBP on BSJ (Exon and Intron)
Plot
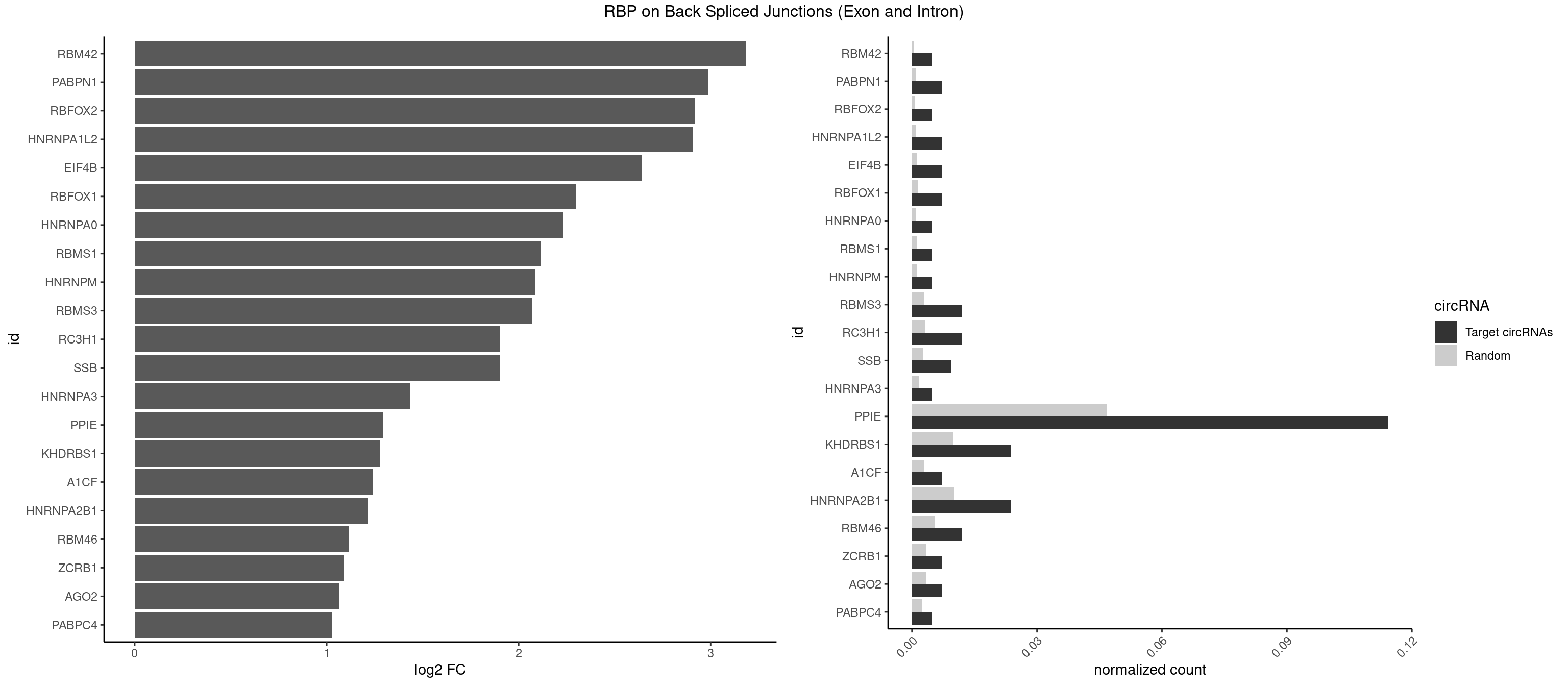
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA | AAAAGA,AGAAGA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGU,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| RBMS3 | 4 | 562 | 0.011904762 | 0.0028365578 | 2.069326 | AAUAUA,AUAUAG,CAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RC3H1 | 4 | 631 | 0.011904762 | 0.0031841999 | 1.902536 | CUUCUG,UCCCUU,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| PPIE | 47 | 9262 | 0.114285714 | 0.0466696896 | 1.292087 | AAAAUA,AAAUAU,AAUAAU,AAUAUA,AAUUUU,AUAAAA,AUAAUA,AUAAUU,AUAUAA,AUAUUU,AUUAAU,AUUUAA,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAUAA,UAAUUU,UAUAAA,UAUAAU,UAUAUU,UAUUAA,UAUUUU,UUAAAA,UUAAUA,UUAUAA,UUAUAU,UUAUUU,UUUAAA,UUUAUA,UUUUAA,UUUUAU,UUUUUA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| KHDRBS1 | 9 | 1945 | 0.023809524 | 0.0098045143 | 1.280021 | AUAAAA,AUUUAA,CUAAAA,UAAAAA,UAAAAC,UAAAAG,UUAAAA | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPA2B1 | 9 | 2033 | 0.023809524 | 0.0102478839 | 1.216213 | AAGGAA,AAGGAG,AGAUAU,AUUUAA,UAGGGU,UUAGGG,UUUAUA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AUGAUA,AUGAUU,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AUUUAA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.