circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000075218:+:22:46326435:46326654
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000075218:+:22:46326435:46326654 | ENSG00000075218 | MSTRG.22339.2 | + | 22 | 46326436 | 46326654 | 219 | GGUCACUGUCCACAGCACCCCGGUUAGACGCUCAUCUGGGCCAGCACCACAAAGCCUGCUGAGCGCAUGGCGUGUGUCAGCCUUGCCCACACCCGCCAGCCGGCGCUGCUCUGGCCUUCCACCGAUGACCCCCAAAACGAUGCCCAGGGCCGUGGGCUCUCCCCUGUGUGUGCCAGCUCGGAGACGUUCCUCUGAGCCCCGCAAGAACUCUGCAAUGAGGGUCACUGUCCACAGCACCCCGGUUAGACGCUCAUCUGGGCCAGCACCAC | circ |
| ENSG00000075218:+:22:46326435:46326654 | ENSG00000075218 | MSTRG.22339.2 | + | 22 | 46326436 | 46326654 | 22 | UCUGCAAUGAGGGUCACUGUCC | bsj |
| ENSG00000075218:+:22:46326435:46326654 | ENSG00000075218 | MSTRG.22339.2 | + | 22 | 46326236 | 46326445 | 210 | GCCAGGAUGGGUUGGUAGCAGCUCGGAGCCUUGCCCGCCUUGGGGCCUGUCCCAGGAGAGCGGAGCUCACACGGGCUCCCUGGCUUGGCUCUUCUGGGCUGAAACCAAAGCCCUCAGCACUGCAUUAGCACAGGCUGAAUGAUGAGAUCUUACUUUUUCUAUGUCAUCUCAGCUCACGACACUGAUGUUGUGUUUGCCAGGGUCACUGUC | ie_up |
| ENSG00000075218:+:22:46326435:46326654 | ENSG00000075218 | MSTRG.22339.2 | + | 22 | 46326645 | 46326854 | 210 | CUGCAAUGAGGUAAGACACGAAGGUGGUAAUAAAUUGGUCUCAAAUUCCUAGGCUUGCUGUCCCAGACAGUGACAUACCUUUUUCUUGCCUUGCUCCAGGUUUUAGUGAAGUAAAUAGUAUUUGGAAAGUUUGUUUGUUUUUUUUUUCAUUGCAAAGAGAAAAUGCAAUAGACAUGAGCAUAGAGGAAAAUUAAAGGUAUUACAAGGAAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
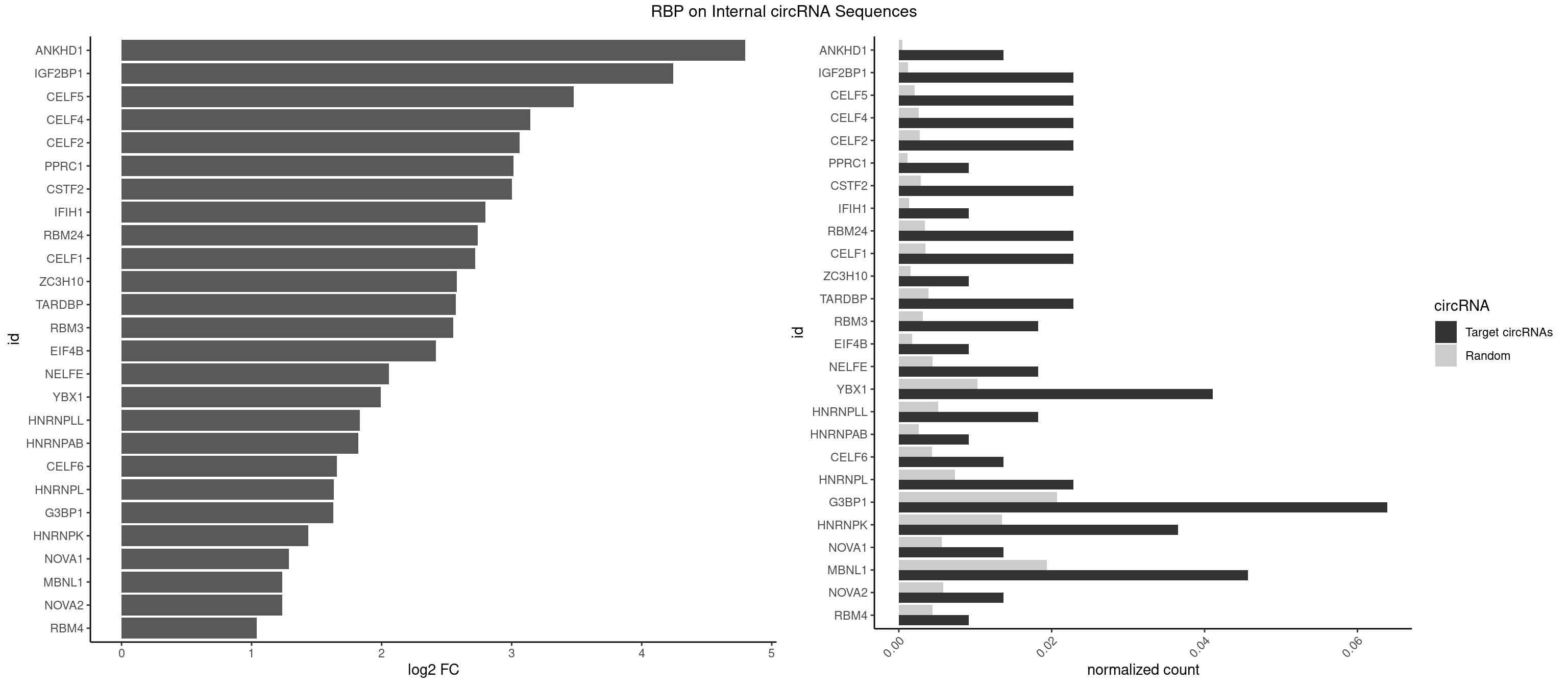
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 339 | 0.01369863 | 0.0004927293 | 4.797092 | AGACGU,GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| IGF2BP1 | 4 | 831 | 0.02283105 | 0.0012057377 | 4.243009 | AGCACC,CACCCG,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| CELF5 | 4 | 1415 | 0.02283105 | 0.0020520728 | 3.475843 | GUGUGU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 4 | 1782 | 0.02283105 | 0.0025839306 | 3.143358 | GUGUGU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF2 | 4 | 1886 | 0.02283105 | 0.0027346479 | 3.061570 | GUGUGU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PPRC1 | 1 | 780 | 0.00913242 | 0.0011318283 | 3.012342 | CGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| CSTF2 | 4 | 1967 | 0.02283105 | 0.0028520334 | 3.000934 | GUGUGU,UGUGUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| IFIH1 | 1 | 904 | 0.00913242 | 0.0013115296 | 2.799747 | GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM24 | 4 | 2357 | 0.02283105 | 0.0034172229 | 2.740101 | GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF1 | 4 | 2391 | 0.02283105 | 0.0034664959 | 2.719447 | GUGUGU,UGUGUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ZC3H10 | 1 | 1053 | 0.00913242 | 0.0015274610 | 2.579862 | GAGCGC | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| TARDBP | 4 | 2654 | 0.02283105 | 0.0038476365 | 2.568953 | GUGUGU,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBM3 | 3 | 2152 | 0.01826484 | 0.0031201361 | 2.549388 | AAAACG,AAACGA,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| EIF4B | 1 | 1179 | 0.00913242 | 0.0017100607 | 2.416950 | CUCGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| NELFE | 3 | 3028 | 0.01826484 | 0.0043896388 | 2.056895 | CUCUGG,GGUUAG,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| YBX1 | 8 | 7119 | 0.04109589 | 0.0103183321 | 1.993784 | ACCACA,CACACC,CACCAC,CAUCUG,CCACAA,CCACAC,CCAGCA,GCCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| HNRNPLL | 3 | 3534 | 0.01826484 | 0.0051229360 | 1.834026 | ACCACA,CACACC,CACCAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| HNRNPAB | 1 | 1782 | 0.00913242 | 0.0025839306 | 1.821430 | ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CELF6 | 2 | 2997 | 0.01369863 | 0.0043447134 | 1.656699 | UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| HNRNPL | 4 | 5085 | 0.02283105 | 0.0073706513 | 1.631133 | ACCACA,CACAAA,CACACC,CACCAC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| G3BP1 | 13 | 14282 | 0.06392694 | 0.0206989801 | 1.626864 | ACCCCC,ACCCGC,CACCCG,CCACAC,CCACAG,CCACCG,CCCACA,CCCAGG,CCCCCA,CCCCGC,CCCCGG,CCCGCA,CCCGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| HNRNPK | 7 | 9304 | 0.03652968 | 0.0134848428 | 1.437730 | ACCCCC,ACCCGC,CCCCAA,CCCCCA,GCCCAC,GCCCAG,UCCCCU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| NOVA1 | 2 | 3872 | 0.01369863 | 0.0056127669 | 1.287248 | AGCACC | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| MBNL1 | 9 | 13372 | 0.04566210 | 0.0193802045 | 1.236413 | ACGCUC,AUGCCC,CCUGCU,CGCUGC,CUGCUC,CUGCUG,GCGCUG,GCUGCU,UUGCCC | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| NOVA2 | 2 | 4013 | 0.01369863 | 0.0058171047 | 1.235658 | AGCACC | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| RBM4 | 1 | 3065 | 0.00913242 | 0.0044432593 | 1.039379 | CCUUCC | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
RBP on BSJ (Exon Only)
Plot
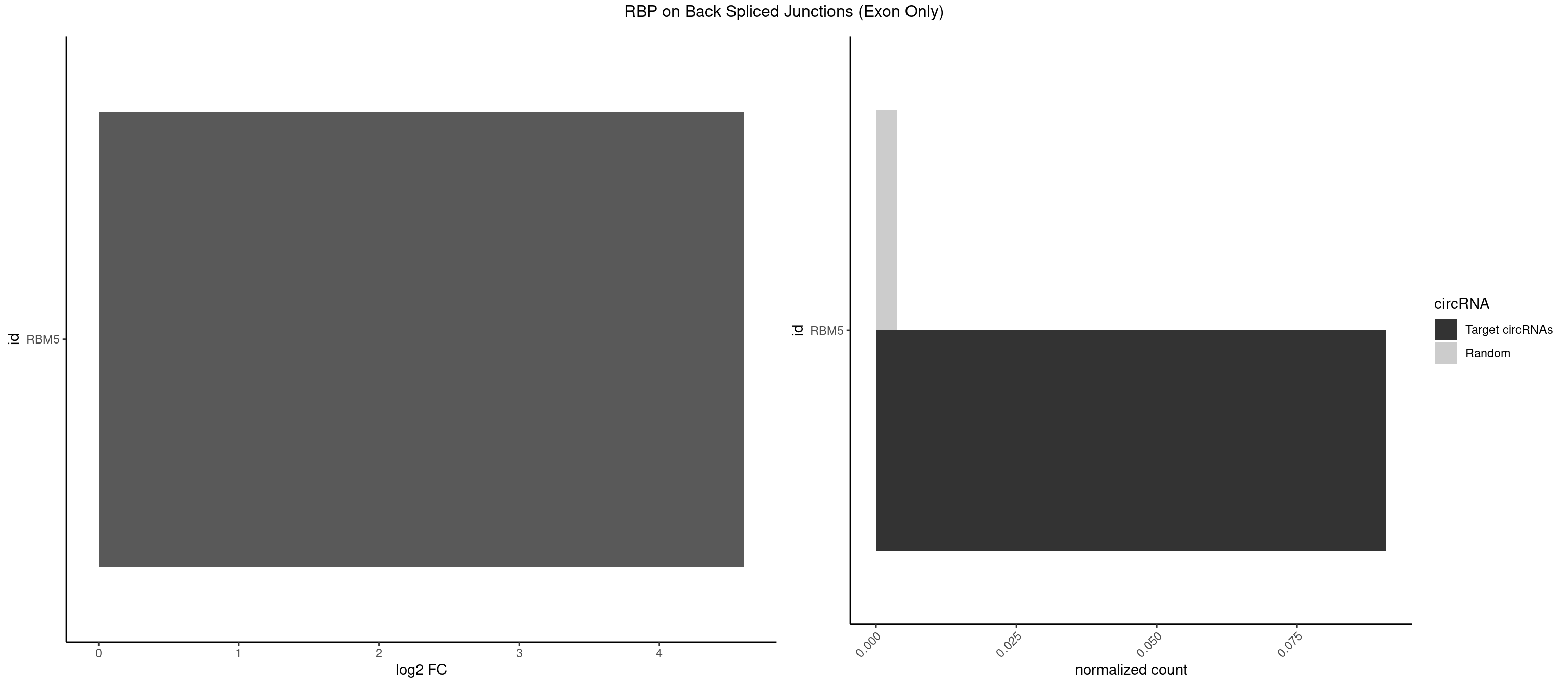
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM5 | 1 | 56 | 0.09090909 | 0.003733298 | 4.605902 | GAGGGU | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
RBP on BSJ (Exon and Intron)
Plot
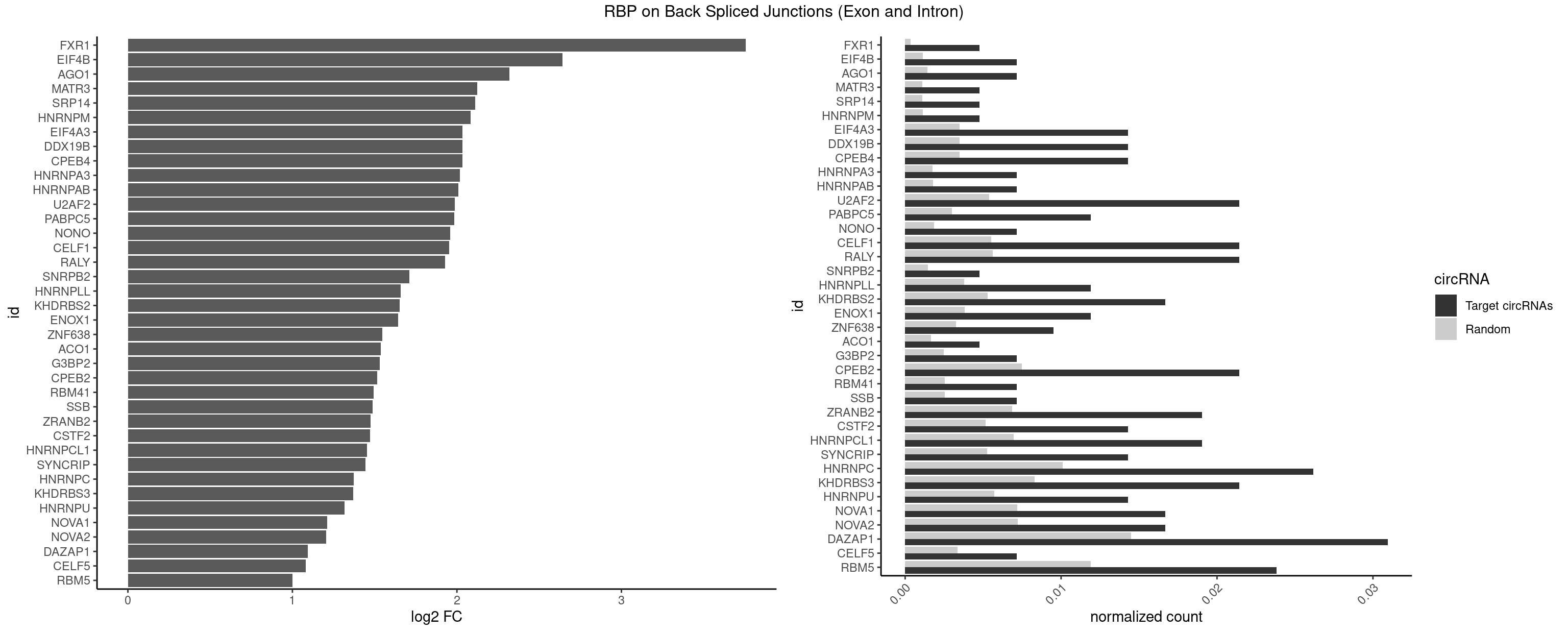
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | ACGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUCGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | GAGGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| CPEB4 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| DDX19B | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 5 | 691 | 0.014285714 | 0.0034864974 | 2.034723 | UUUUUU | UUUUUU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | CAAGGA,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AAGACA,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| U2AF2 | 8 | 1071 | 0.021428571 | 0.0054010480 | 1.988224 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| PABPC5 | 4 | 596 | 0.011904762 | 0.0030078597 | 1.984730 | AGAAAA,GAAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| NONO | 2 | 364 | 0.007142857 | 0.0018389762 | 1.957598 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| CELF1 | 8 | 1097 | 0.021428571 | 0.0055320435 | 1.953651 | GUUGUG,GUUUGU,UGUGUU,UGUUGU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| RALY | 8 | 1117 | 0.021428571 | 0.0056328094 | 1.927609 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | AUUGCA | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | ACAUAC,ACGACA,ACUGCA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| KHDRBS2 | 6 | 1051 | 0.016666667 | 0.0053002821 | 1.652825 | AAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| ENOX1 | 4 | 756 | 0.011904762 | 0.0038139863 | 1.642167 | AAGACA,AGACAG,CAGACA,UAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUGG,GUUGGU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| CPEB2 | 8 | 1487 | 0.021428571 | 0.0074969770 | 1.515155 | CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ZRANB2 | 7 | 1360 | 0.019047619 | 0.0068571141 | 1.473937 | AAAGGU,AGGUAA,AGGUAU,AGGUUU,GGUGGU,UAAAGG,UGGUAA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.0051541717 | 1.470761 | GUGUUU,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| HNRNPCL1 | 7 | 1381 | 0.019047619 | 0.0069629182 | 1.451847 | CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SYNCRIP | 5 | 1041 | 0.014285714 | 0.0052498992 | 1.444212 | UUUUUU | AAAAAA,UUUUUU |
| HNRNPC | 10 | 2006 | 0.026190476 | 0.0101118501 | 1.372995 | CUUUUU,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS3 | 8 | 1646 | 0.021428571 | 0.0082980653 | 1.368689 | AAUAAA,AUUAAA,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPU | 5 | 1136 | 0.014285714 | 0.0057285369 | 1.318335 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| NOVA1 | 6 | 1428 | 0.016666667 | 0.0071997179 | 1.210953 | UUCAUU,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| NOVA2 | 6 | 1434 | 0.016666667 | 0.0072299476 | 1.204908 | AGACAU,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| DAZAP1 | 12 | 2878 | 0.030952381 | 0.0145052398 | 1.093476 | AGGAAA,AGGAUG,AGGUAA,AGUAAA,AGUUUG,UAGUAU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM5 | 9 | 2359 | 0.023809524 | 0.0118903668 | 1.001746 | AAGGAA,AAGGUA,AAGGUG,AGGUAA,CAAGGA,CUCUUC,GAAGGU,GGUGGU,UCUUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.