circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000169855:-:3:78639743:78647655
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000169855:-:3:78639743:78647655 | ENSG00000169855 | ENST00000436010 | - | 3 | 78639744 | 78647655 | 225 | UCCCGUCUUUUACCUUCACACCAACAGUAACUUACCAGAGAGGAGGCGAAGCUGUCAGCAGUGGAGGGAGGCCUGGACUUCUCAACAUCAGUGAACCUGCCGCGCAGCCAUGGCUGGCAGACACGUGGCCUAAUACUGGCAACAACCACAAUGACUGCUCCAUCAGCUGCUGCACGGCAGGCAAUGGAAACAGCGACAGCAACCUCACUACCUACAGUCGCCCAGUCCCGUCUUUUACCUUCACACCAACAGUAACUUACCAGAGAGGAGGCGAA | circ |
| ENSG00000169855:-:3:78639743:78647655 | ENSG00000169855 | ENST00000436010 | - | 3 | 78639744 | 78647655 | 22 | CAGUCGCCCAGUCCCGUCUUUU | bsj |
| ENSG00000169855:-:3:78639743:78647655 | ENSG00000169855 | ENST00000436010 | - | 3 | 78647646 | 78647855 | 210 | UCAAAAUGGUACCUGUAGAGACAAGCUUCUUUUUUUAACCUCAGAAUUGGUUUUAUUCAUAGAUCAGAUUGUCAGAACUACUGGCUUAGUUAUUUACAUUUGUUUUAUGUUCAGCUGUCUGCUUGAUUUAUACUAAUGUGAUAUGUUCCCUUUCUCUCUUCAGCUUAAUAACUGGUUUAUAUUUUGUUUUGAUUUUUCAGUCCCGUCUUU | ie_up |
| ENSG00000169855:-:3:78639743:78647655 | ENSG00000169855 | ENST00000436010 | - | 3 | 78639544 | 78639753 | 210 | AGUCGCCCAGGUUAGGGACACAGAGAGAGCCUGAUAUGUAUAAGCUUUUUAAGGAACUAGCCAGAAGAUCUUUUCUUUCCCUCUCCCUACAUGGGAAAGAUAAAAUUUGACCCGUAUUUUAUCCAAUGCUGACUAUUGUUUAGCAAAAUAGCCUGAUUUAAGUAUUUUCUCCAUCCCUUAACCUCCAUACUUUGCUAAAGUCCACCAAAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
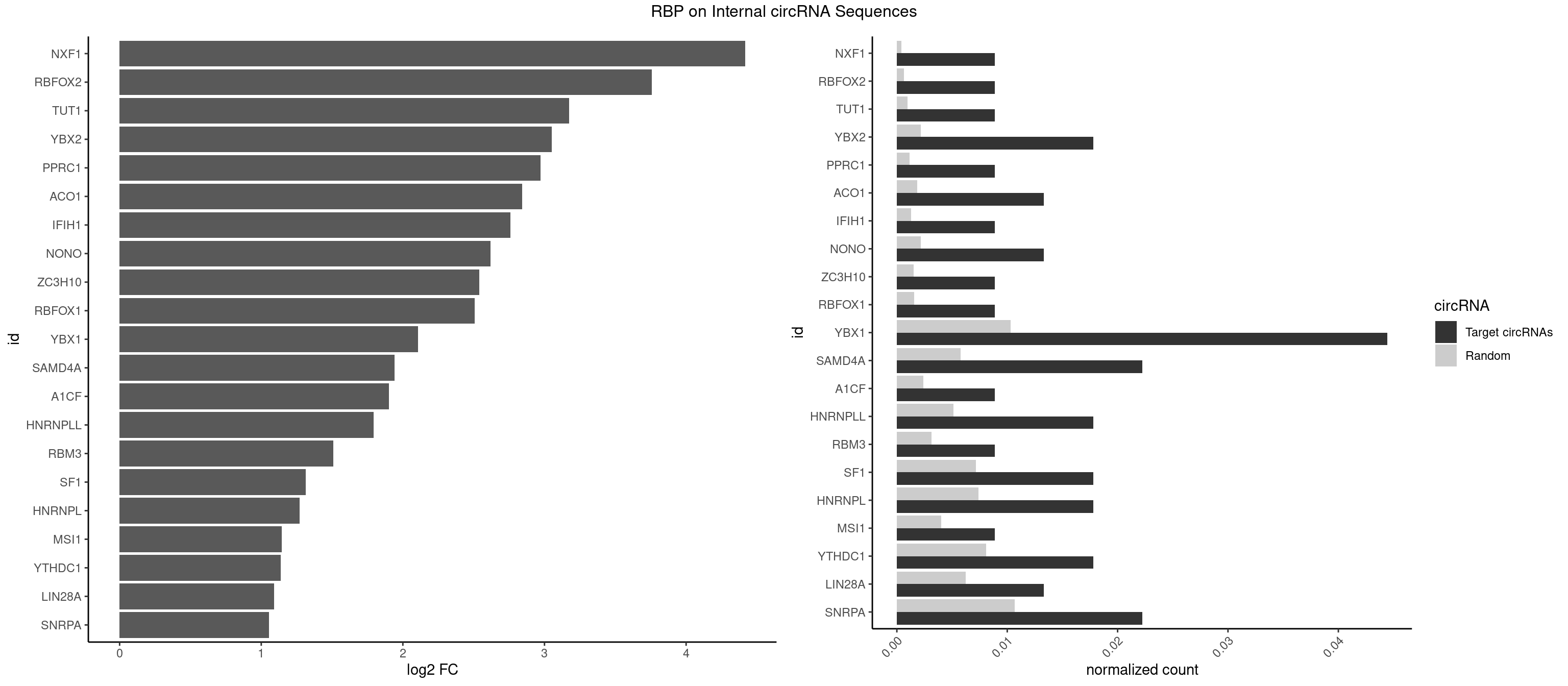
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 286 | 0.008888889 | 0.0004159215 | 4.417620 | AACCUG | AACCUG |
| RBFOX2 | 1 | 452 | 0.008888889 | 0.0006564894 | 3.759159 | UGACUG | UGACUG,UGCAUG |
| TUT1 | 1 | 678 | 0.008888889 | 0.0009840095 | 3.175259 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| YBX2 | 3 | 1480 | 0.017777778 | 0.0021462711 | 3.050171 | AACAAC,AACAUC,ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PPRC1 | 1 | 780 | 0.008888889 | 0.0011318283 | 2.973348 | CCGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| ACO1 | 2 | 1283 | 0.013333333 | 0.0018607779 | 2.841060 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| IFIH1 | 1 | 904 | 0.008888889 | 0.0013115296 | 2.760753 | GCCGCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| NONO | 2 | 1498 | 0.013333333 | 0.0021723567 | 2.617705 | AGAGGA,GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ZC3H10 | 1 | 1053 | 0.008888889 | 0.0015274610 | 2.540868 | CAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBFOX1 | 1 | 1077 | 0.008888889 | 0.0015622419 | 2.508385 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| YBX1 | 9 | 7119 | 0.044444444 | 0.0103183321 | 2.106793 | AACAUC,AACCAC,ACACCA,ACAUCA,ACCACA,CAACCA,CACACC,CAGCAA,CCACAA | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| SAMD4A | 4 | 3992 | 0.022222222 | 0.0057866714 | 1.941197 | CUGGAC,CUGGCA,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| A1CF | 1 | 1642 | 0.008888889 | 0.0023810421 | 1.900410 | AUCAGU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPLL | 3 | 3534 | 0.017777778 | 0.0051229360 | 1.795032 | ACACCA,ACCACA,CACACC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBM3 | 1 | 2152 | 0.008888889 | 0.0031201361 | 1.510394 | AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SF1 | 3 | 4931 | 0.017777778 | 0.0071474739 | 1.314570 | ACAGUC,AGUAAC,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| HNRNPL | 3 | 5085 | 0.017777778 | 0.0073706513 | 1.270211 | ACACCA,ACCACA,CACACC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| MSI1 | 1 | 2770 | 0.008888889 | 0.0040157442 | 1.146336 | AGGAGG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| YTHDC1 | 3 | 5576 | 0.017777778 | 0.0080822104 | 1.137253 | GACUGC,UAAUAC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| LIN28A | 2 | 4315 | 0.013333333 | 0.0062547643 | 1.092010 | GGAGGG,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SNRPA | 4 | 7380 | 0.022222222 | 0.0106965744 | 1.054854 | ACCUGC,AGCAGU,UGCACG,UUACCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
RBP on BSJ (Exon Only)
Plot
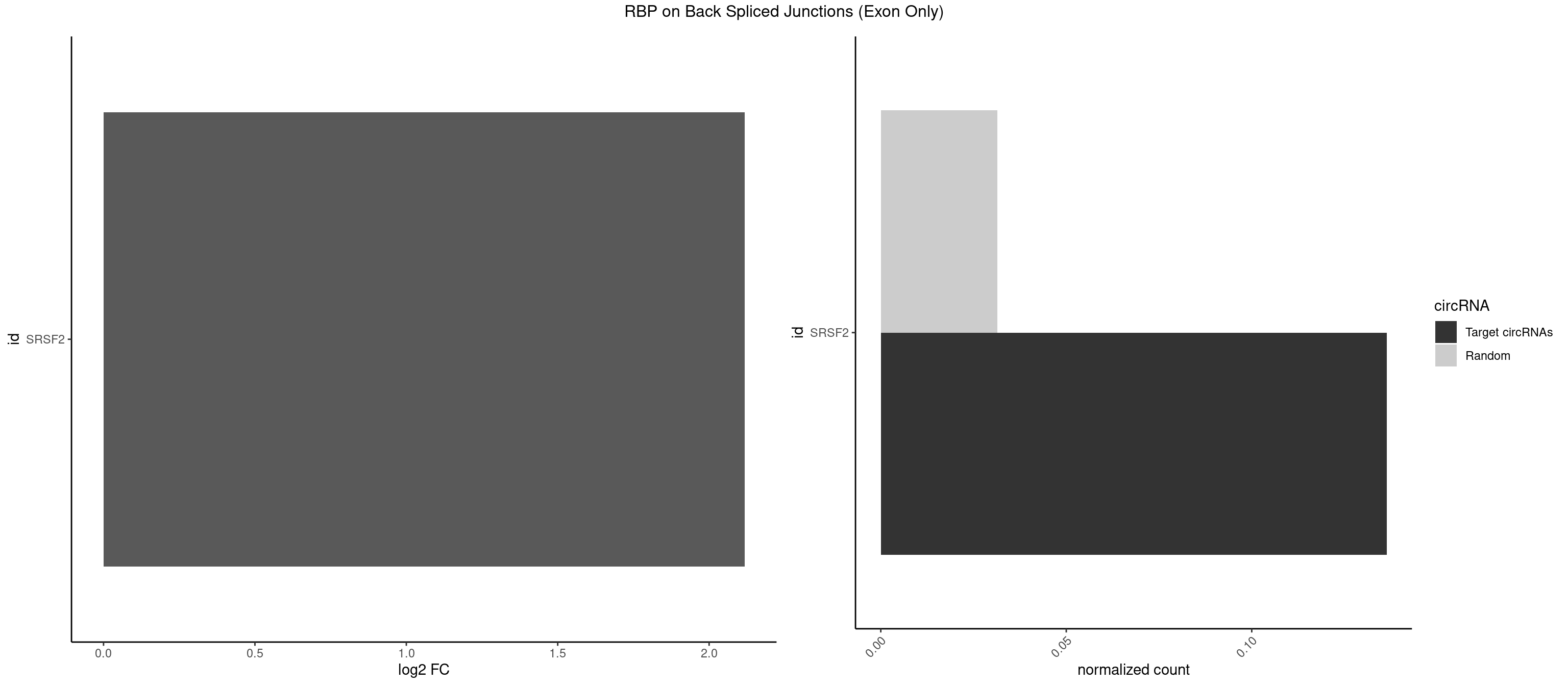
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF2 | 2 | 479 | 0.1363636 | 0.0314383 | 2.116864 | CAGUCC,CCAGUC | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
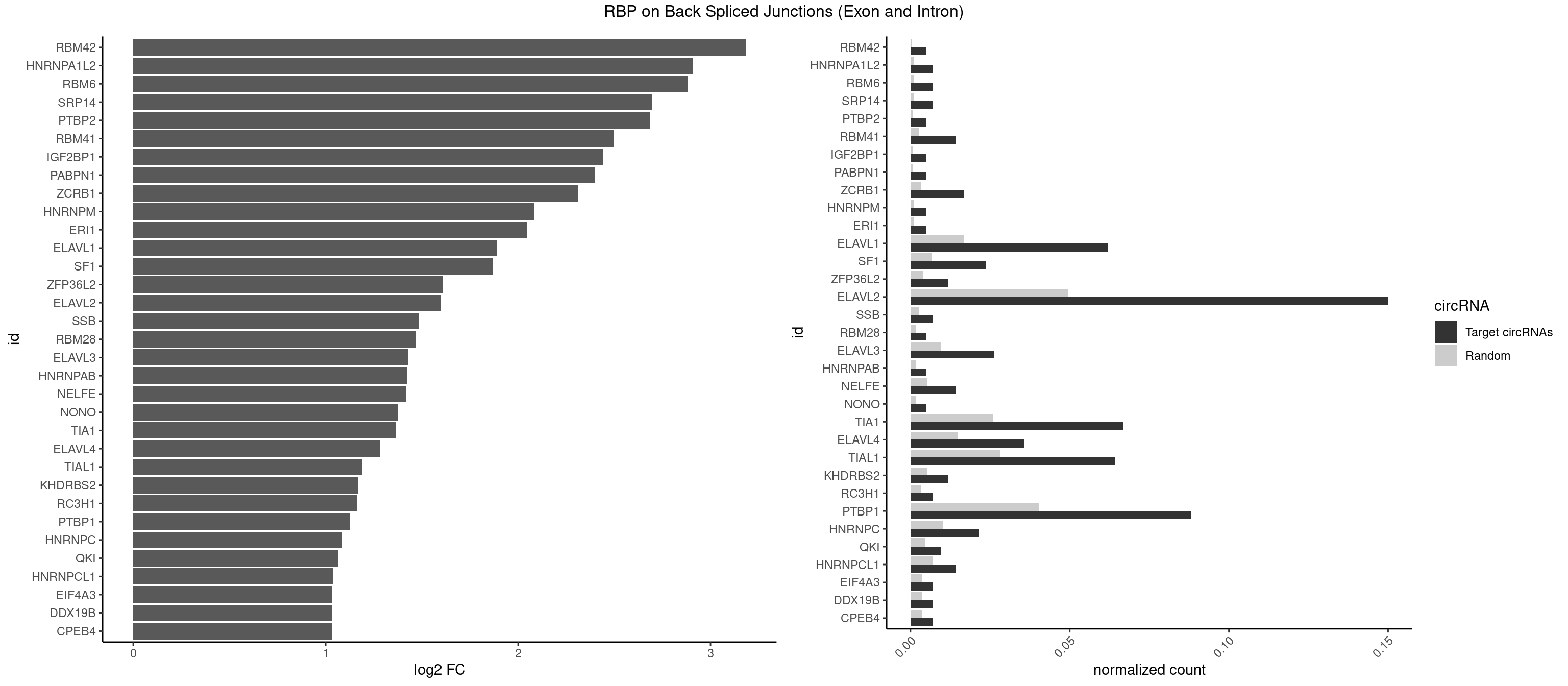
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AUCCAA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CCUGUA,CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| RBM41 | 5 | 502 | 0.014285714 | 0.0025342604 | 2.494937 | AUACUU,UACAUG,UACAUU,UACUUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | ACCCGU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| ZCRB1 | 6 | 666 | 0.016666667 | 0.0033605401 | 2.310201 | AUUUAA,GAUUUA,GCUUAA,GGCUUA,GGUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| ELAVL1 | 25 | 3309 | 0.061904762 | 0.0166767432 | 1.892213 | AUUUAU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAUU,UUUGUU,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| SF1 | 9 | 1294 | 0.023809524 | 0.0065245869 | 1.867580 | ACUAAU,ACUAGC,AUACUA,CACAGA,GCUGAC,UACUAA,UAUACU,UGCUAA,UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| ELAVL2 | 62 | 9826 | 0.150000000 | 0.0495112858 | 1.599133 | AAUUUG,AUACUU,AUAUUU,AUUUAA,AUUUAC,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CUUUCU,CUUUUU,GAUUUA,GAUUUU,GUUUUA,UACUUU,UAGUUA,UAUACU,UAUAUU,UAUGUU,UAUUGU,UAUUUA,UAUUUU,UCUUUU,UGAUUU,UUAAGU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUU,UUCUUU,UUGAUU,UUUAAG,UUUAUA,UUUAUG,UUUAUU,UUUCUU,UUUGAU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ELAVL3 | 10 | 1928 | 0.026190476 | 0.0097188634 | 1.430183 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NELFE | 5 | 1059 | 0.014285714 | 0.0053405885 | 1.419503 | CUCUCU,CUGGCU,CUGGUU,GGUUAG,UCUCUC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| TIA1 | 27 | 5140 | 0.066666667 | 0.0259018541 | 1.363910 | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUUUUC,CUUUUU,GUUUUA,GUUUUG,UAUUUU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCU,UUUUGU,UUUUUA,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL4 | 14 | 2916 | 0.035714286 | 0.0146966949 | 1.281010 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| TIAL1 | 26 | 5597 | 0.064285714 | 0.0282043531 | 1.188580 | AAAUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CUUUUC,CUUUUU,UAUUUU,UUAUUU,UUCAGU,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | AUAAAA,GAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PTBP1 | 36 | 8003 | 0.088095238 | 0.0403264813 | 1.127337 | AGCUGU,AUUUUC,AUUUUU,CCUUUC,CUCCAU,CUCUCU,CUCUUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GUCUUU,UACUUU,UAUUUU,UCCAUC,UCUCUC,UCUCUU,UCUUUC,UCUUUU,UUAUUU,UUCUCU,UUCUUU,UUUCCC,UUUCUU,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| HNRNPC | 8 | 2006 | 0.021428571 | 0.0101118501 | 1.083489 | AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | ACUAAU,UAACCU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.0069629182 | 1.036809 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.