circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000163468:-:1:156333546:156335888
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000163468:-:1:156333546:156335888 | ENSG00000163468 | ENST00000368262 | - | 1 | 156333547 | 156335888 | 273 | GCCAGAACACAAAGCGUGAAUCCGGAAGAAAAGUUCAAUCUGGAAACAUCAAUGCUGCCAAGACUAUUGCAGAUAUCAUCCGAACAUGUUUGGGACCCAAGUCCAUGAUGAAGAUGCUUUUGGACCCAAUGGGAGGCAUUGUGAUGACCAAUGAUGGCAAUGCCAUUCUUCGAGAGAUUCAAGUCCAGCAUCCAGCGGCCAAGUCCAUGAUCGAAAUUAGCCGGACCCAGGAUGAAGAGGUUGGAGAUGGGACCACAUCAGUAAUUAUUCUUGGCCAGAACACAAAGCGUGAAUCCGGAAGAAAAGUUCAAUCUGGAAACAUC | circ |
| ENSG00000163468:-:1:156333546:156335888 | ENSG00000163468 | ENST00000368262 | - | 1 | 156333547 | 156335888 | 22 | AAUUAUUCUUGGCCAGAACACA | bsj |
| ENSG00000163468:-:1:156333546:156335888 | ENSG00000163468 | ENST00000368262 | - | 1 | 156335879 | 156336088 | 210 | UAAGGACAUAUGUUUACAGUUUUUUGAGAUAUGGUUGACAUAUAAUAAACUGCACUUAUUUAAAGUGUAGAGUUUGAUAAGCUUUGCUCUGUGGAUAUACCAUGAAACCUGCAUUCUUAUUUUUCAAAGACGGGGAAAUAAAUAGCUUGUGUACGAUGGGGCAGUCCUGGGCUGUUGACUUGUGUUUUUGUCUAUUUUAGGCCAGAACAC | ie_up |
| ENSG00000163468:-:1:156333546:156335888 | ENSG00000163468 | ENST00000368262 | - | 1 | 156333347 | 156333556 | 210 | AUUAUUCUUGGUAAGAGGAGAAUAAGAGGUUGAUAAGGAAAAAUUAGAAGUAACACCUGUGUCAUAUUUUAAGAAGGAAAUUCUGUUACAGAUCCACUAAUUACAGAUAUUUAAAUAGCUUAUUGGUUAUUUUCUAUGUAUUCUCUCCAGUUGGGUAUAAAACAUUUAUAUGAAUAGGGUGAGCUAUGCAAAUCUGGCUGAUAAACUAAU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
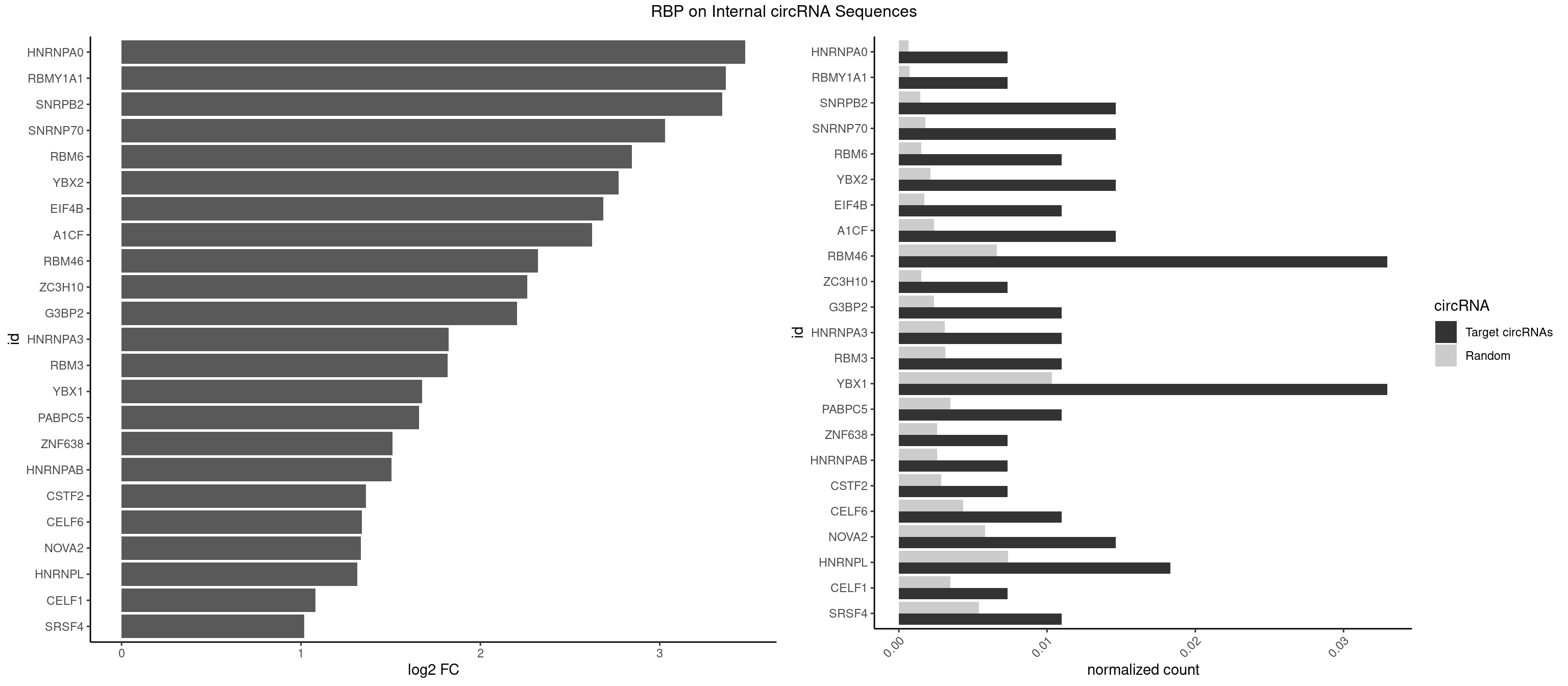
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 1 | 453 | 0.007326007 | 0.0006579386 | 3.477002 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMY1A1 | 1 | 489 | 0.007326007 | 0.0007101099 | 3.366913 | CAAGAC | ACAAGA,CAAGAC |
| SNRPB2 | 3 | 991 | 0.014652015 | 0.0014376103 | 3.349354 | AUUGCA,UAUUGC,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| SNRNP70 | 3 | 1237 | 0.014652015 | 0.0017941145 | 3.029755 | AUUCAA,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM6 | 2 | 1054 | 0.010989011 | 0.0015289102 | 2.845486 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| YBX2 | 3 | 1480 | 0.014652015 | 0.0021462711 | 2.771195 | AACAUC,ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| EIF4B | 2 | 1179 | 0.010989011 | 0.0017100607 | 2.683942 | GUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| A1CF | 3 | 1642 | 0.014652015 | 0.0023810421 | 2.621434 | AUCAGU,UAAUUA,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBM46 | 8 | 4554 | 0.032967033 | 0.0066011240 | 2.320240 | AAUGAU,AUCAAU,AUGAAG,AUGAUG,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ZC3H10 | 1 | 1053 | 0.007326007 | 0.0015274610 | 2.261892 | CCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| G3BP2 | 2 | 1644 | 0.010989011 | 0.0023839405 | 2.204641 | AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPA3 | 2 | 2140 | 0.010989011 | 0.0031027457 | 1.824444 | GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM3 | 2 | 2152 | 0.010989011 | 0.0031201361 | 1.816381 | AAGACU,AGACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| YBX1 | 8 | 7119 | 0.032967033 | 0.0103183321 | 1.675814 | AACAUC,ACAUCA,ACCACA,AUCAUC,CCAGCA,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| PABPC5 | 2 | 2400 | 0.010989011 | 0.0034795387 | 1.659094 | AGAAAA,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZNF638 | 1 | 1773 | 0.007326007 | 0.0025708878 | 1.510760 | GGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPAB | 1 | 1782 | 0.007326007 | 0.0025839306 | 1.503460 | ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CSTF2 | 1 | 1967 | 0.007326007 | 0.0028520334 | 1.361036 | UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF6 | 2 | 2997 | 0.010989011 | 0.0043447134 | 1.338729 | GUGAUG,UGUGAU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| NOVA2 | 3 | 4013 | 0.014652015 | 0.0058171047 | 1.332726 | AGGCAU,AUCAUC,GAGGCA | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| HNRNPL | 4 | 5085 | 0.018315018 | 0.0073706513 | 1.313163 | AACACA,ACACAA,ACCACA,CACAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| CELF1 | 1 | 2391 | 0.007326007 | 0.0034664959 | 1.079549 | UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SRSF4 | 2 | 3740 | 0.010989011 | 0.0054214720 | 1.019305 | GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
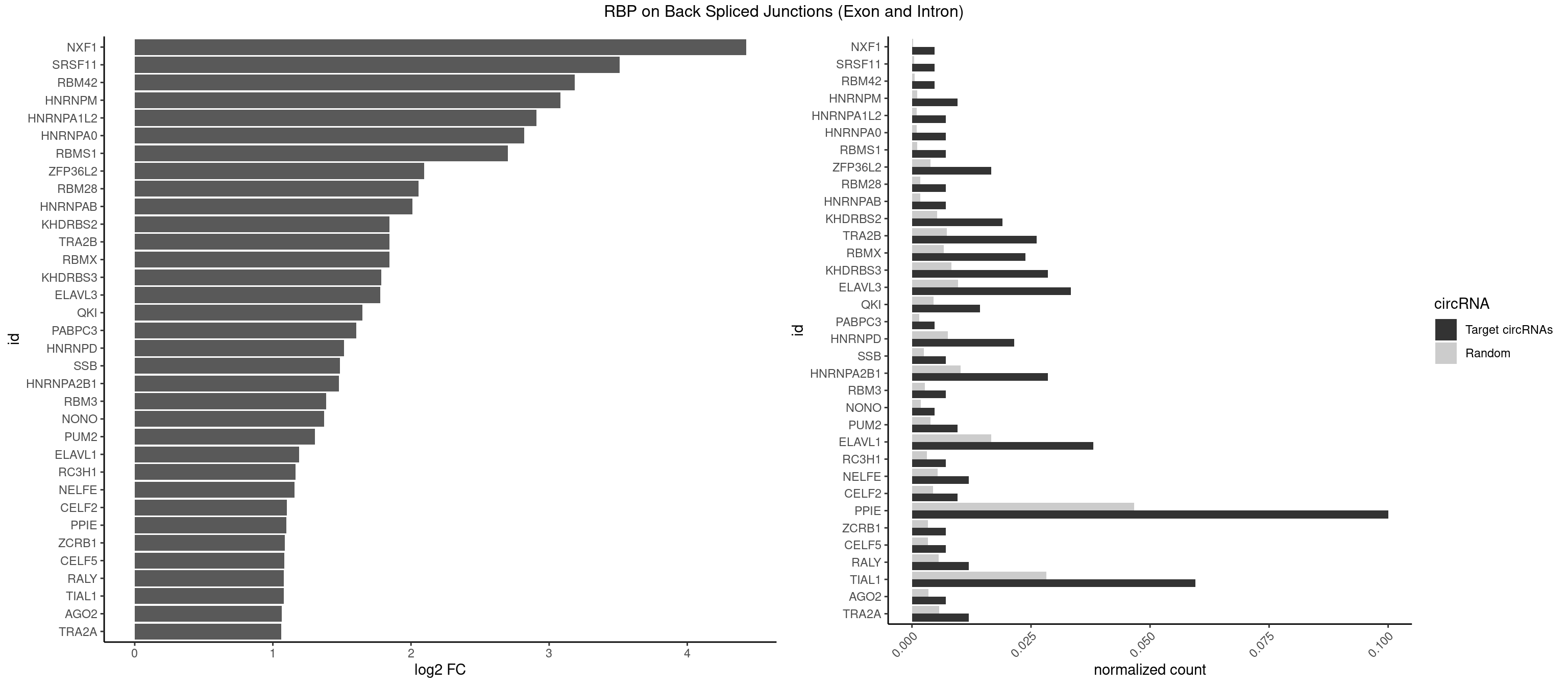
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| HNRNPM | 3 | 222 | 0.009523810 | 0.0011235389 | 3.083489 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | AUAGGG,UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMS1 | 2 | 217 | 0.007142857 | 0.0010983474 | 2.701167 | AUAUAC,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| ZFP36L2 | 6 | 774 | 0.016666667 | 0.0039046755 | 2.093691 | AUUUAU,UAUUUA,UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | GUGUAG,UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AAAGAC,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| KHDRBS2 | 7 | 1051 | 0.019047619 | 0.0053002821 | 1.845470 | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| TRA2B | 10 | 1447 | 0.026190476 | 0.0072954454 | 1.843974 | AAGAAG,AAGGAA,AAUAAG,AGAAGG,AGGAAA,AUAAGA,GAAGGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBMX | 9 | 1316 | 0.023809524 | 0.0066354293 | 1.843277 | AAGAAG,AAGGAA,AAGUAA,AAGUGU,AGAAGG,AGUAAC,GAAGGA,GUAACA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| KHDRBS3 | 11 | 1646 | 0.028571429 | 0.0082980653 | 1.783726 | AAAUAA,AAUAAA,AUAAAA,AUAAAC,GAUAAA,UAAAAC,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ELAVL3 | 13 | 1928 | 0.033333333 | 0.0097188634 | 1.778106 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | ACUAAU,ACUUAU,CACUAA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPD | 8 | 1488 | 0.021428571 | 0.0075020153 | 1.514186 | AGAUAU,AUUUAU,UAUUUA,UUAUUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPA2B1 | 11 | 2033 | 0.028571429 | 0.0102478839 | 1.479247 | AAGAAG,AAGGAA,AGAUAU,AUAGGG,AUUUAA,GAAGGA,UAGGGU,UUUAUA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAACUA,AAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UAAAUA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ELAVL1 | 15 | 3309 | 0.038095238 | 0.0166767432 | 1.191773 | AUUUAU,UAUUUA,UAUUUU,UGUUUU,UUAUUU,UUGGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUGGCU,UCUCUC,UCUGGC,UGGUUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | UAUGUU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PPIE | 41 | 9262 | 0.100000000 | 0.0466696896 | 1.099442 | AAAAAU,AAAAUU,AAAUAA,AAAUUA,AAUAAA,AUAAAA,AUAAAU,AUAAUA,AUAUAA,AUAUUU,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAUA,UAAUAA,UAAUUA,UAUAAA,UAUAAU,UAUUUA,UAUUUU,UUAAAU,UUAUAU,UUAUUU,UUUAAA,UUUAUA,UUUUAA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| TIAL1 | 24 | 5597 | 0.059523810 | 0.0282043531 | 1.077549 | AGUUUU,AUUUUA,AUUUUC,AUUUUU,CAGUUU,GUUUUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUUAA,UUUUCA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AAGAAG,AAGAGG,AGAGGA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.