circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000112290:-:6:110127468:110127629
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000112290:-:6:110127468:110127629 | ENSG00000112290 | ENST00000392588 | - | 6 | 110127469 | 110127629 | 161 | GUGAACUGGCACAAAGGUUAAUCUCAAGAUGCCGCUAGUGAAAAGAAACAUCGAUCCUAGGCACUUGUGCCACACAGCACUGCCUAGAGGCAUUAAGAAUGAACUGGAAUGUGUAACCAAUAUUUCCUUGGCAAAUAUAAUUAGACAACUAAGUAGCCUAAGUGAACUGGCACAAAGGUUAAUCUCAAGAUGCCGCUAGUGAAAAGAAACA | circ |
| ENSG00000112290:-:6:110127468:110127629 | ENSG00000112290 | ENST00000392588 | - | 6 | 110127469 | 110127629 | 22 | AAGUAGCCUAAGUGAACUGGCA | bsj |
| ENSG00000112290:-:6:110127468:110127629 | ENSG00000112290 | ENST00000392588 | - | 6 | 110127620 | 110127829 | 210 | GGCUGAGGAGGAAGAGGAGGGGUUUGUCCUGCUGUCUCAGGGAGGCAGAGAGGGAAGAAAAUCCACCAGUAAGUGGACCCAGGCAGUUCAAAUCCAUGUUGUUCAAGGGUCAACAGUGCUUUAUUCUUCAAUGAAAAAUAAAGUCUCAUUCUGUGUUGGCUGAAAUUUAAUAUUGUUAAUUUAUUUUUGAUUUCCUACAGGUGAACUGGC | ie_up |
| ENSG00000112290:-:6:110127468:110127629 | ENSG00000112290 | ENST00000392588 | - | 6 | 110127269 | 110127478 | 210 | AGUAGCCUAAGUAAGUAUAUAUCAAUUAAAAAUAUACUCACAAACCAGAAUGUUAUGCUUUAGCGUAUUAAGAAGUUGUGCUUCUGAUUAAUUUGGGAGUAUGAUUUGGUGAGAAGAGUACAAAAUUAUCCUGAAAAAUAUAGUCACAUGAGUAUAAUUAUUGAGUUUUACAAGAACCUCUUCUGCAUGAUGUUUCUUCUGGGAAAGGAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
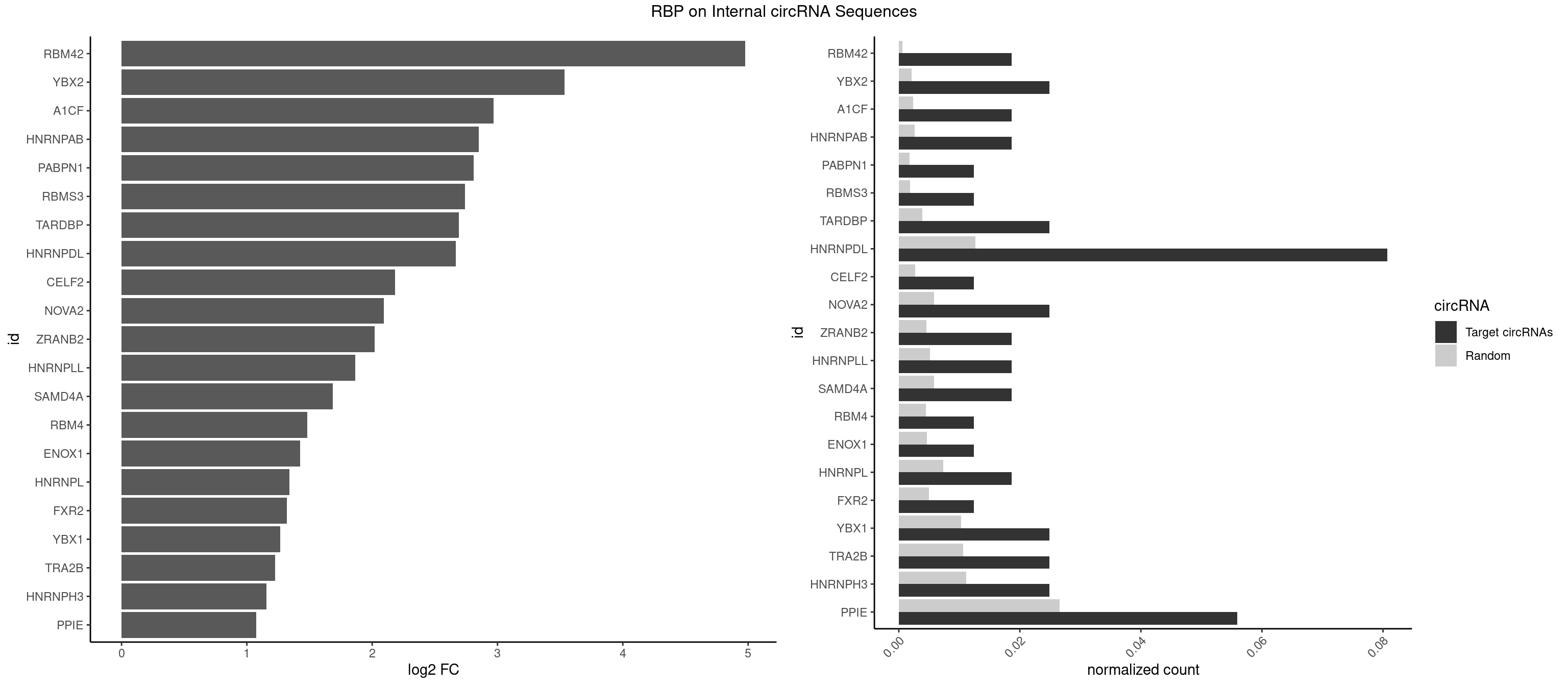
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 2 | 407 | 0.01863354 | 0.0005912752 | 4.977928 | AACUAA,ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| YBX2 | 3 | 1480 | 0.02484472 | 0.0021462711 | 3.533035 | AACAUC,ACAACU,ACAUCG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| A1CF | 2 | 1642 | 0.01863354 | 0.0023810421 | 2.968237 | AUAAUU,UAAUUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPAB | 2 | 1782 | 0.01863354 | 0.0025839306 | 2.850263 | ACAAAG,AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPN1 | 1 | 1222 | 0.01242236 | 0.0017723764 | 2.809182 | AAAAGA | AAAAGA,AGAAGA |
| RBMS3 | 1 | 1283 | 0.01242236 | 0.0018607779 | 2.738962 | AAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| TARDBP | 3 | 2654 | 0.02484472 | 0.0038476365 | 2.690895 | GAAUGA,GAAUGU,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| HNRNPDL | 12 | 8759 | 0.08074534 | 0.0126950266 | 2.669116 | AACUAA,ACAGCA,ACUAAG,AGUAGC,CUAAGU,CUAGAG,CUAGGC,GCUAGU,GUAGCC,UAAGUA,UAGGCA | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| CELF2 | 1 | 1886 | 0.01242236 | 0.0027346479 | 2.183512 | AUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| NOVA2 | 3 | 4013 | 0.02484472 | 0.0058171047 | 2.094566 | AGGCAU,CCUAGA,GAGGCA | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| ZRANB2 | 2 | 3173 | 0.01863354 | 0.0045997733 | 2.018267 | AAAGGU,AGGUUA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| HNRNPLL | 2 | 3534 | 0.01863354 | 0.0051229360 | 1.862859 | CACACA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| SAMD4A | 2 | 3992 | 0.01863354 | 0.0057866714 | 1.687096 | CUGGAA,CUGGCA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| RBM4 | 1 | 3065 | 0.01242236 | 0.0044432593 | 1.483249 | UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ENOX1 | 1 | 3195 | 0.01242236 | 0.0046316558 | 1.423339 | UAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPL | 2 | 5085 | 0.01863354 | 0.0073706513 | 1.338038 | CACAAA,CACACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| FXR2 | 1 | 3434 | 0.01242236 | 0.0049780156 | 1.319297 | AGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| YBX1 | 3 | 7119 | 0.02484472 | 0.0103183321 | 1.267730 | AACAUC,ACAUCG,CCACAC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| TRA2B | 3 | 7329 | 0.02484472 | 0.0106226650 | 1.225794 | AAAGAA,AAGAAU,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| HNRNPH3 | 3 | 7688 | 0.02484472 | 0.0111429292 | 1.156811 | AAUGUG,GAAUGU,GGAAUG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| PPIE | 8 | 18315 | 0.05590062 | 0.0265436196 | 1.074499 | AAAUAU,AAUAUA,AAUAUU,AUAAUU,AUAUAA,AUAUUU,UAAUUA,UAUAAU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
RBP on BSJ (Exon Only)
Plot
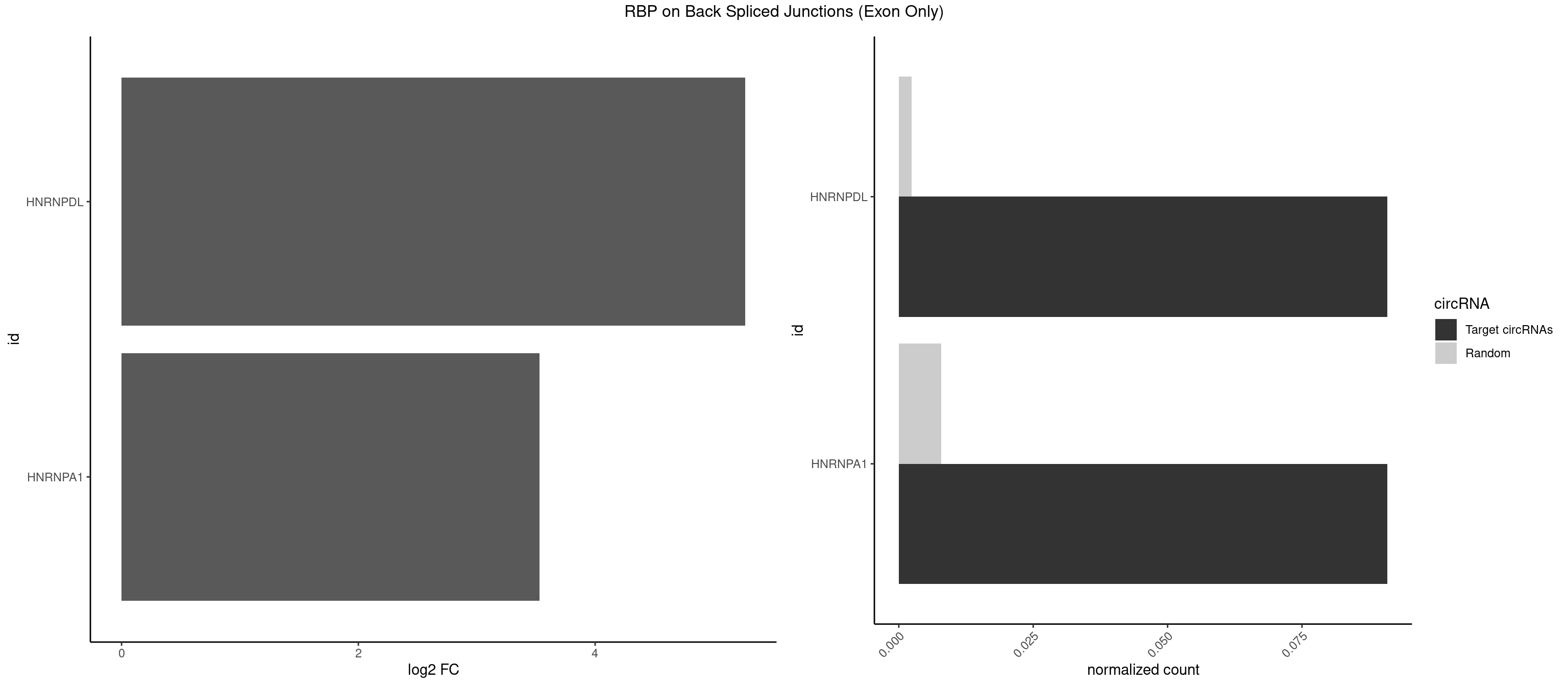
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPDL | 1 | 35 | 0.09090909 | 0.002357873 | 5.268867 | CUAAGU | AACAGC,AAUACC,AAUUUA,ACACCA,ACAGCA,ACCAGA,AGAUAU,AGUAGG,AUCUGA,AUUAGC,AUUAGG,CACGCA,CCAGAC,CUAAGC,CUAAGU,CUAGAU,CUAGGA,CUAGGC,CUUUAG,GAACUA,GAUUAG,GCACUA,GCUAGU,UGCGCA,UUAGCC,UUAGGC,UUUAGG |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.007859576 | 3.531901 | AGUGAA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
RBP on BSJ (Exon and Intron)
Plot
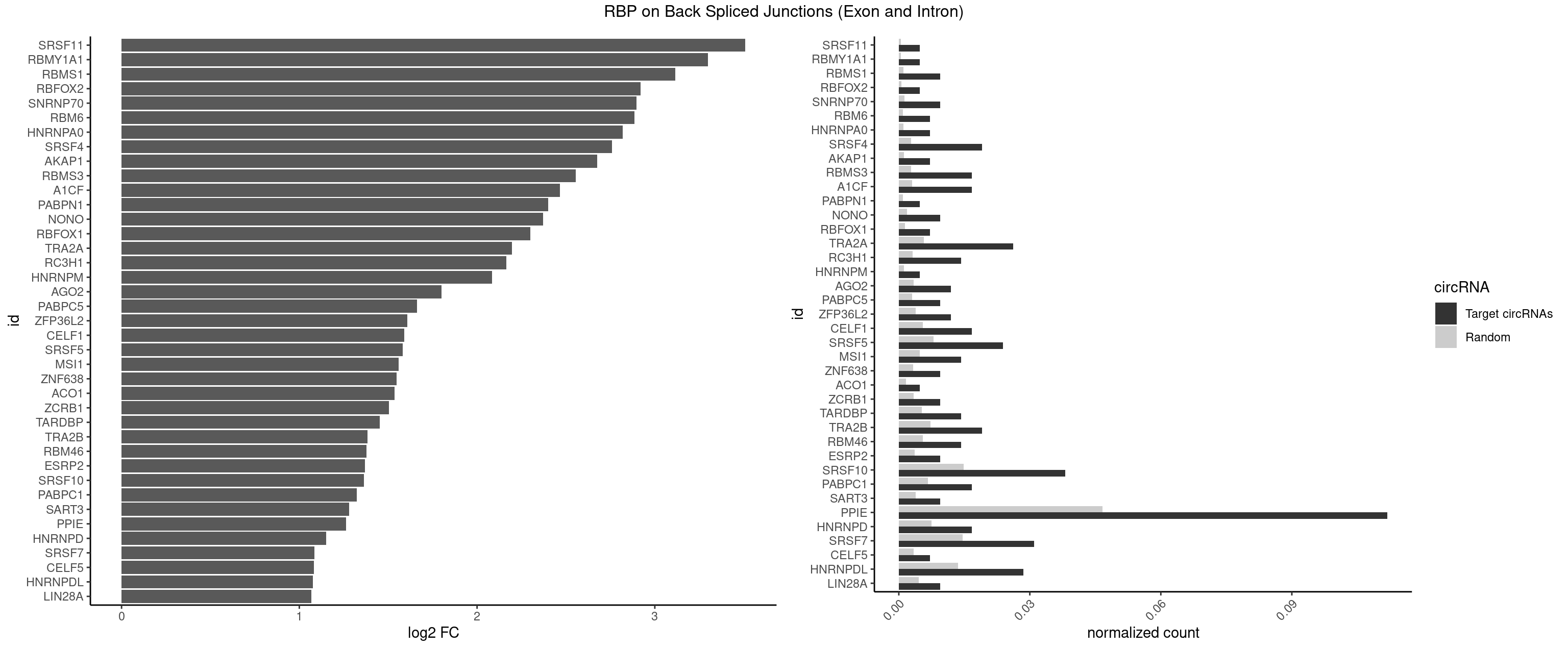
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | ACAAGA | ACAAGA,CAAGAC |
| RBMS1 | 3 | 217 | 0.009523810 | 0.0010983474 | 3.116204 | AUAUAC,AUAUAG,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| SNRNP70 | 3 | 253 | 0.009523810 | 0.0012797259 | 2.895704 | GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| SRSF4 | 7 | 558 | 0.019047619 | 0.0028164047 | 2.757684 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| RBMS3 | 6 | 562 | 0.016666667 | 0.0028365578 | 2.554752 | AAUAUA,AUAUAG,AUAUAU,UAUAUA,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| A1CF | 6 | 598 | 0.016666667 | 0.0030179363 | 2.465331 | AGUAUA,AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| NONO | 3 | 364 | 0.009523810 | 0.0018389762 | 2.372636 | AGAGGA,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGA,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| TRA2A | 10 | 1133 | 0.026190476 | 0.0057134220 | 2.196615 | AAGAAA,AAGAAG,AAGAGG,AGAAGA,AGAGGA,AGGAAG,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RC3H1 | 5 | 631 | 0.014285714 | 0.0031841999 | 2.165570 | CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| AGO2 | 4 | 677 | 0.011904762 | 0.0034159613 | 1.801175 | AGUGCU,GUGCUU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAA,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | AUUUAU,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| CELF1 | 6 | 1097 | 0.016666667 | 0.0055320435 | 1.591081 | CUGUCU,GUGUUG,GUUGUG,GUUUGU,UGUGUU,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SRSF5 | 9 | 1578 | 0.023809524 | 0.0079554615 | 1.581521 | AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,GAAGAA,GAGGAA,GGAAGA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| MSI1 | 5 | 961 | 0.014285714 | 0.0048468360 | 1.559458 | AGGAAG,AGGAGG,AGUAAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GUUGUU,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AAUUAA,AUUUAA,GAUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| TARDBP | 5 | 1034 | 0.014285714 | 0.0052146312 | 1.453936 | GAAUGU,GUUGUG,GUUGUU,UUGUGC,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| TRA2B | 7 | 1447 | 0.019047619 | 0.0072954454 | 1.384543 | AAGAAC,AAGAAG,AAGGAA,AGAAGA,AGGAAG,GAAGAA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBM46 | 5 | 1091 | 0.014285714 | 0.0055018138 | 1.376594 | AAUGAA,AUCAAU,AUGAAA,AUGAUG,AUGAUU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGAAG,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SRSF10 | 15 | 2937 | 0.038095238 | 0.0148024990 | 1.363770 | AAAGGA,AAGAAA,AAGAAG,AAGAGG,AAGGAA,AGAGAG,AGAGGA,AGAGGG,GAGAAG,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| PABPC1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | ACAAAC,AGAAAA,CAAACC,CAAAUC,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PPIE | 46 | 9262 | 0.111904762 | 0.0466696896 | 1.261714 | AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AUAAUU,AUAUAU,AUUAAA,AUUAAU,AUUAUU,AUUUAA,AUUUAU,AUUUUU,UAAAAA,UAAUAU,UAAUUA,UAAUUU,UAUAAU,UAUAUA,UAUUAA,UAUUUU,UUAAAA,UUAAUA,UUAAUU,UUAUUU,UUUAAU,UUUAUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPD | 6 | 1488 | 0.016666667 | 0.0075020153 | 1.151615 | AAUUUA,AUUUAU,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| SRSF7 | 12 | 2893 | 0.030952381 | 0.0145808142 | 1.085979 | AAAGGA,AAGAAG,AGAAGA,AGAGAG,AGAGGA,AGGAAG,CAGAGA,CUCUUC,GAAGAA,GAGGAA,UCAACA,UCUUCA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPDL | 11 | 2689 | 0.028571429 | 0.0135530028 | 1.075961 | AAUUUA,ACCAGA,AGUAGC,CACCAG,CUAAGU,CUUUAG,GGAGUA,GUAGCC,UAAGUA | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | GGAGGA,GGAGGG,GGAGUA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.