circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000156639:+:6:38061592:38116739
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000156639:+:6:38061592:38116739 | ENSG00000156639 | ENST00000287218 | + | 6 | 38061593 | 38116739 | 417 | AUUUUCAAAAGAAACAGCCAGACGAUGAUUCCGCUCCAAGUACAAGUAACAGCCAAUCAGAUUUGUUUUCCGAAGAGACCACCAGUGACAACAACAAUACCUCGAUAACCACGCCAACUCUUAGUCCCAGCCAGCAGCCGCUUCCGACAGAACUGAAUGUAACUUCACCGAGUAAAGAGGAGUGUGGGCCAUGCACAGACACAGCUCAUGUCUCAUUAAUCACACCAACAAAAAGAUCCUGUGGUACAGAUUCACAGUCUGAGAAUGAGGCUUCACCAGUAAAACGGCCACGACUACUUGAGAAUACGGAACGGUCCGAGGAAACCAGUCGAUCUAAACAGAAGAGUCGACGUCGGUGCUUCCAGUGCCAAACCAAACUGGAGCUGGUGCAGCAGGAAUUGGGAUCGUGUCGCUGCGAUUUUCAAAAGAAACAGCCAGACGAUGAUUCCGCUCCAAGUACAAGUAAC | circ |
| ENSG00000156639:+:6:38061592:38116739 | ENSG00000156639 | ENST00000287218 | + | 6 | 38061593 | 38116739 | 22 | GUGUCGCUGCGAUUUUCAAAAG | bsj |
| ENSG00000156639:+:6:38061592:38116739 | ENSG00000156639 | ENST00000287218 | + | 6 | 38061393 | 38061602 | 210 | UUGGAAAACUUUUAACUCUAGUACCUUCCACUAAUCUUAAACGUUUUUGACCAGUGUUUUACUCCCAUGGUUUUUUAGUCACCCAGAUCAGUCUUUAUUAUUGGUGUUGUUGCAUUUAAUUUUUGUACCAACUUCCAUUGUUUUCUAAUCCUCAGAGACUGUGAACUCCUUAAUUAAGCUCGUUUUCUAUUUUUCUUCAGAUUUUCAAAA | ie_up |
| ENSG00000156639:+:6:38061592:38116739 | ENSG00000156639 | ENST00000287218 | + | 6 | 38116730 | 38116939 | 210 | UGUCGCUGCGGUAAGCAUCUCCCCCAGUGGCGUGAUGGAGACUAUAUCCUUAACACCUUGGCCCAGCUUUGGAAAUUUAAGUGGCUGAAGUCUUUCGUUCUUCUUCUGAGUACUGUAAGUUUGCUACAGUAGUGCAUGCGAUAUAGGUUUGGGGCUUGAGAAAUCAGGAGCAUAGUAUGUUUGCCUCUAAACACCCUUUAUUCUCACACA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
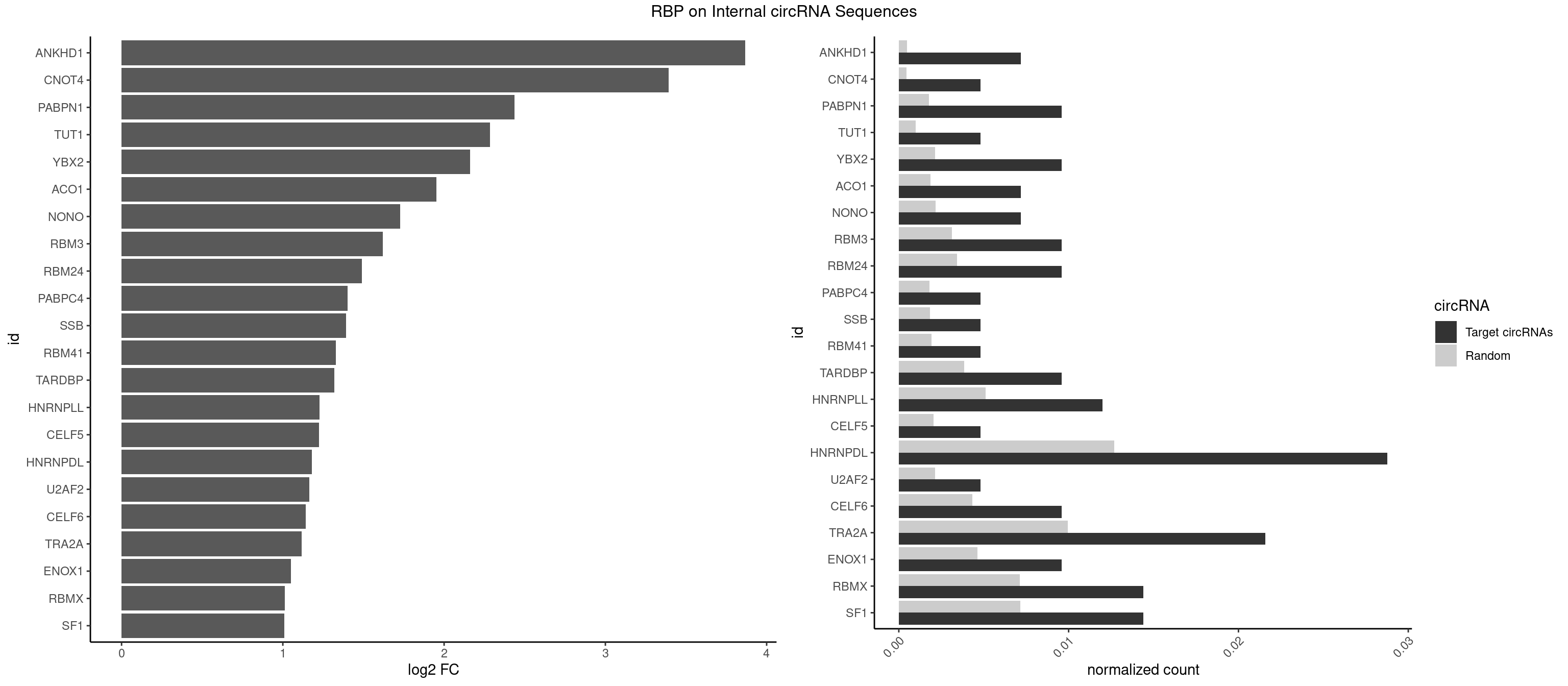
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 339 | 0.007194245 | 0.0004927293 | 3.867976 | AGACGA,GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| CNOT4 | 1 | 314 | 0.004796163 | 0.0004564992 | 3.393196 | GACAGA | GACAGA |
| PABPN1 | 3 | 1222 | 0.009592326 | 0.0017723764 | 2.436196 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| TUT1 | 1 | 678 | 0.004796163 | 0.0009840095 | 2.285137 | CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| YBX2 | 3 | 1480 | 0.009592326 | 0.0021462711 | 2.160048 | AACAAC,ACAACA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| ACO1 | 2 | 1283 | 0.007194245 | 0.0018607779 | 1.950937 | CAGUGA,CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| NONO | 2 | 1498 | 0.007194245 | 0.0021723567 | 1.727582 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM3 | 3 | 2152 | 0.009592326 | 0.0031201361 | 1.620272 | AAAACG,AAUACG,AGACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBM24 | 3 | 2357 | 0.009592326 | 0.0034172229 | 1.489056 | AGUGUG,GAGUGU,GUGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| PABPC4 | 1 | 1251 | 0.004796163 | 0.0018144033 | 1.402386 | AAAAAG | AAAAAA,AAAAAG |
| SSB | 1 | 1260 | 0.004796163 | 0.0018274462 | 1.392052 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM41 | 1 | 1318 | 0.004796163 | 0.0019115000 | 1.327176 | UACUUG | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| TARDBP | 3 | 2654 | 0.009592326 | 0.0038476365 | 1.317908 | GAAUGA,GAAUGU,UGAAUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| HNRNPLL | 4 | 3534 | 0.011990408 | 0.0051229360 | 1.226838 | ACACCA,AGACGA,CACACC,CAGACG | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| CELF5 | 1 | 1415 | 0.004796163 | 0.0020520728 | 1.224799 | GUGUGG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPDL | 11 | 8759 | 0.028776978 | 0.0126950266 | 1.180652 | AACAGC,AAUACC,ACACCA,ACCACG,CACCAG,CCACGC,CCAGAC,UAACAG,UGUCGC | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| U2AF2 | 1 | 1477 | 0.004796163 | 0.0021419234 | 1.162974 | UUUUCC | UUUUCC,UUUUUC,UUUUUU |
| CELF6 | 3 | 2997 | 0.009592326 | 0.0043447134 | 1.142620 | GUGUGG,UGUGGG,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| TRA2A | 8 | 6871 | 0.021582734 | 0.0099589296 | 1.115815 | AAAGAA,AAGAAA,AAGAGG,AGAAGA,AGAGGA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ENOX1 | 3 | 3195 | 0.009592326 | 0.0046316558 | 1.050353 | AGUACA,CAGACA,GUACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RBMX | 5 | 4925 | 0.014388489 | 0.0071387787 | 1.011166 | AAGUAA,ACCAAA,AGUAAC,GUAACA,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SF1 | 5 | 4931 | 0.014388489 | 0.0071474739 | 1.009410 | ACAGAC,ACAGUC,AGUAAC,CACAGA,CACCGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
RBP on BSJ (Exon Only)
Plot
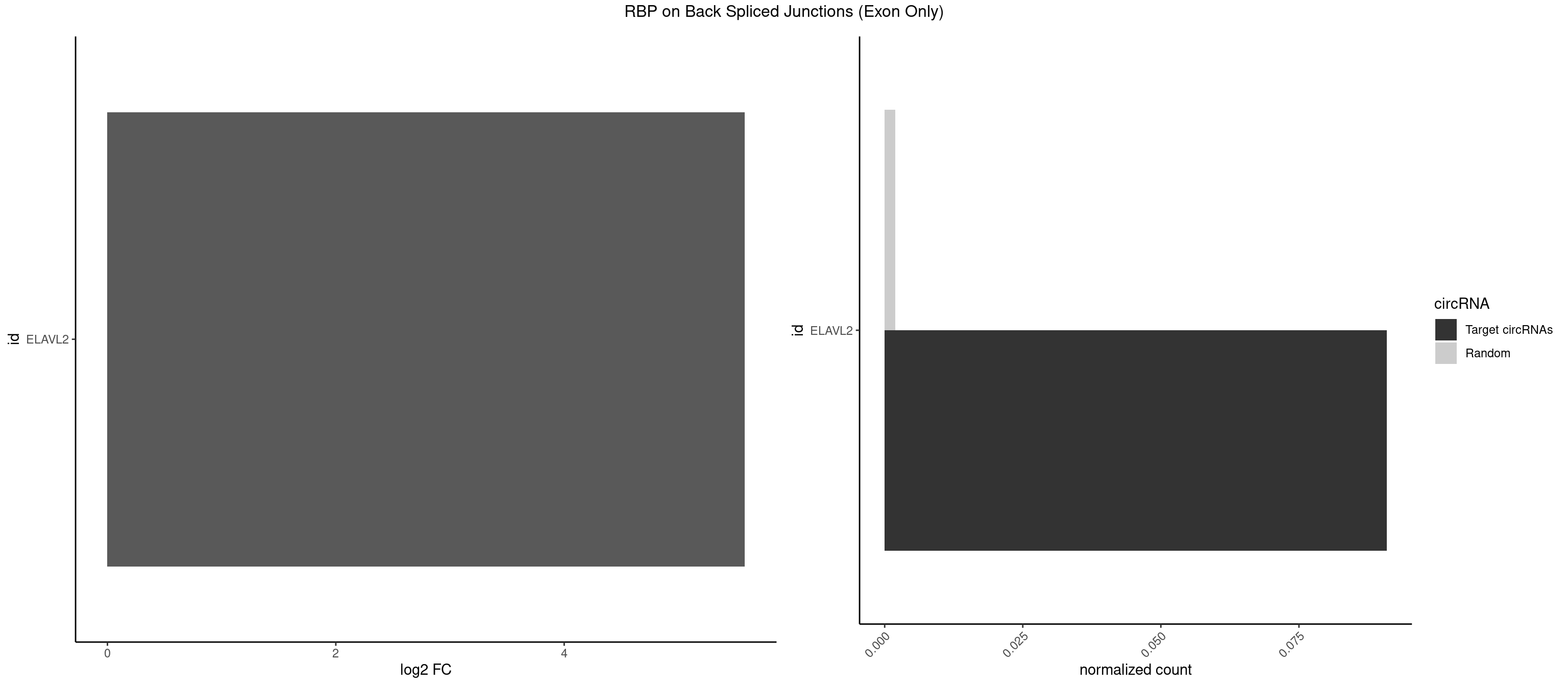
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ELAVL2 | 1 | 28 | 0.09090909 | 0.001899397 | 5.580811 | GAUUUU | AAUUUA,AAUUUU,AUAUUU,AUUUAA,AUUUUG,CUUUCU,GAUUUU,GUAUUG,GUUUUU,UACUUU,UAGUUA,UAUAUA,UAUAUU,UAUGUU,UAUUGA,UAUUUU,UGUAUU,UUAAGU,UUACUU,UUAGUU,UUAUUG,UUUACU,UUUAGU,UUUCUU,UUUUAG |
RBP on BSJ (Exon and Intron)
Plot
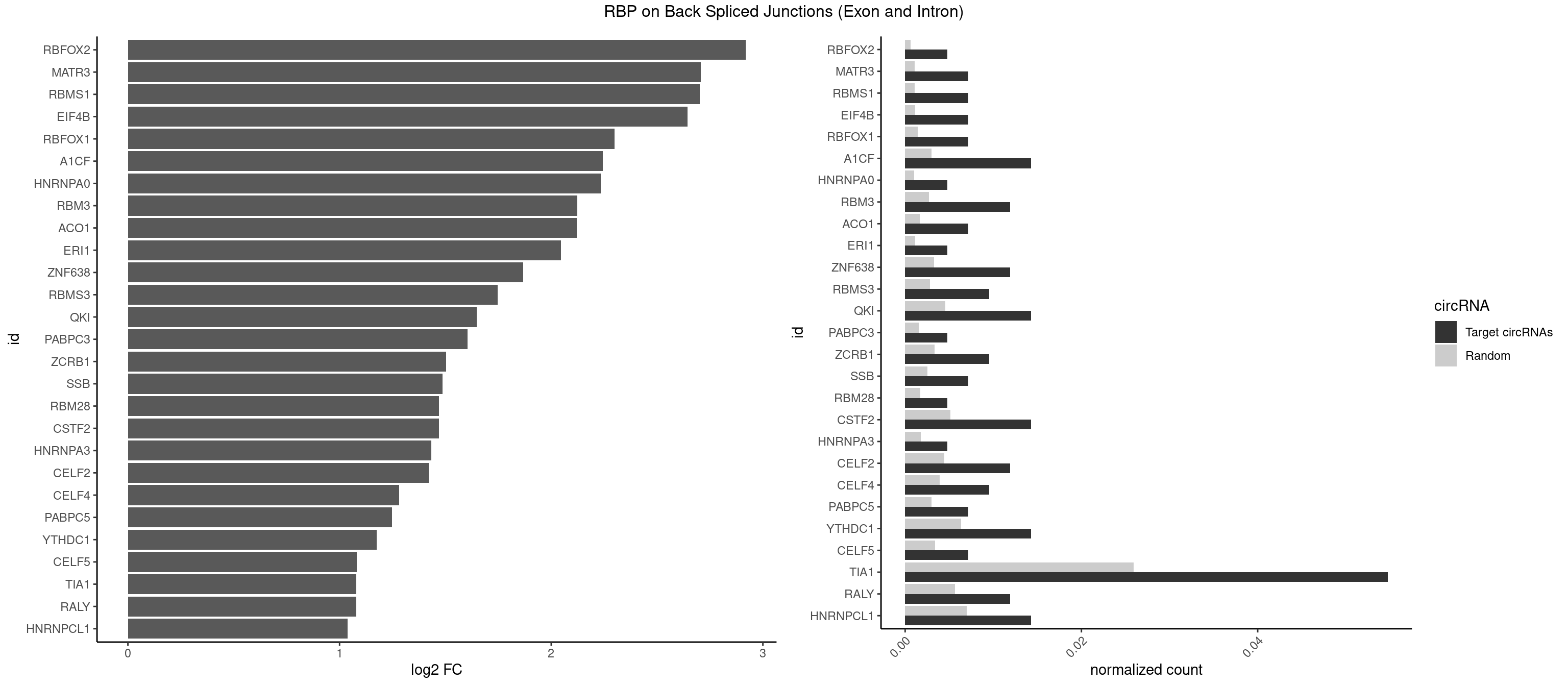
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AAUCUU,AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMS1 | 2 | 217 | 0.007142857 | 0.0010983474 | 2.701167 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGC,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| A1CF | 5 | 598 | 0.014285714 | 0.0030179363 | 2.242939 | AUCAGU,GAUCAG,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| RBM3 | 4 | 541 | 0.011904762 | 0.0027307537 | 2.124168 | AAAACU,AGACUA,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUCAGA | UUCAGA,UUUCAG |
| ZNF638 | 4 | 646 | 0.011904762 | 0.0032597743 | 1.868695 | CGUUCU,GUUCUU,GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAG,CUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | ACUAAU,CACUAA,CUAAUC,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AAUUAA,AUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.0051541717 | 1.470761 | GUGUUG,GUGUUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | AGGAGC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| CELF2 | 4 | 881 | 0.011904762 | 0.0044437727 | 1.421682 | GUAUGU,GUUGUU,UAUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF4 | 3 | 776 | 0.009523810 | 0.0039147521 | 1.282618 | GGUGUU,GUGUUG,GUGUUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GAGUAC,GCAUGC,UAGUAC,UAGUGC,UGCUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUG,GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| TIA1 | 22 | 5140 | 0.054761905 | 0.0259018541 | 1.080117 | AUUUUC,AUUUUU,CUCCUU,CUUUUA,GUUUUA,GUUUUC,GUUUUU,UAUUUU,UUUAUU,UUUUCU,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.0069629182 | 1.036809 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.