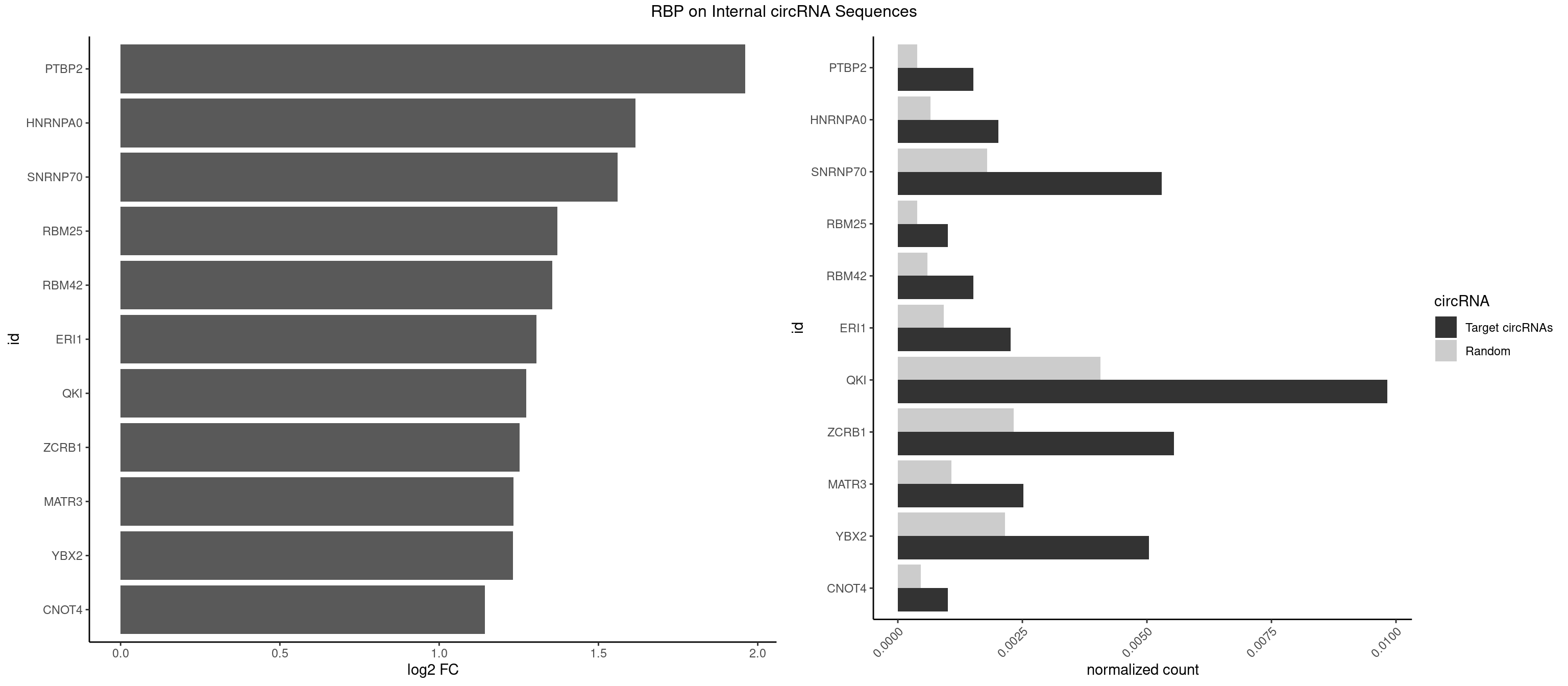
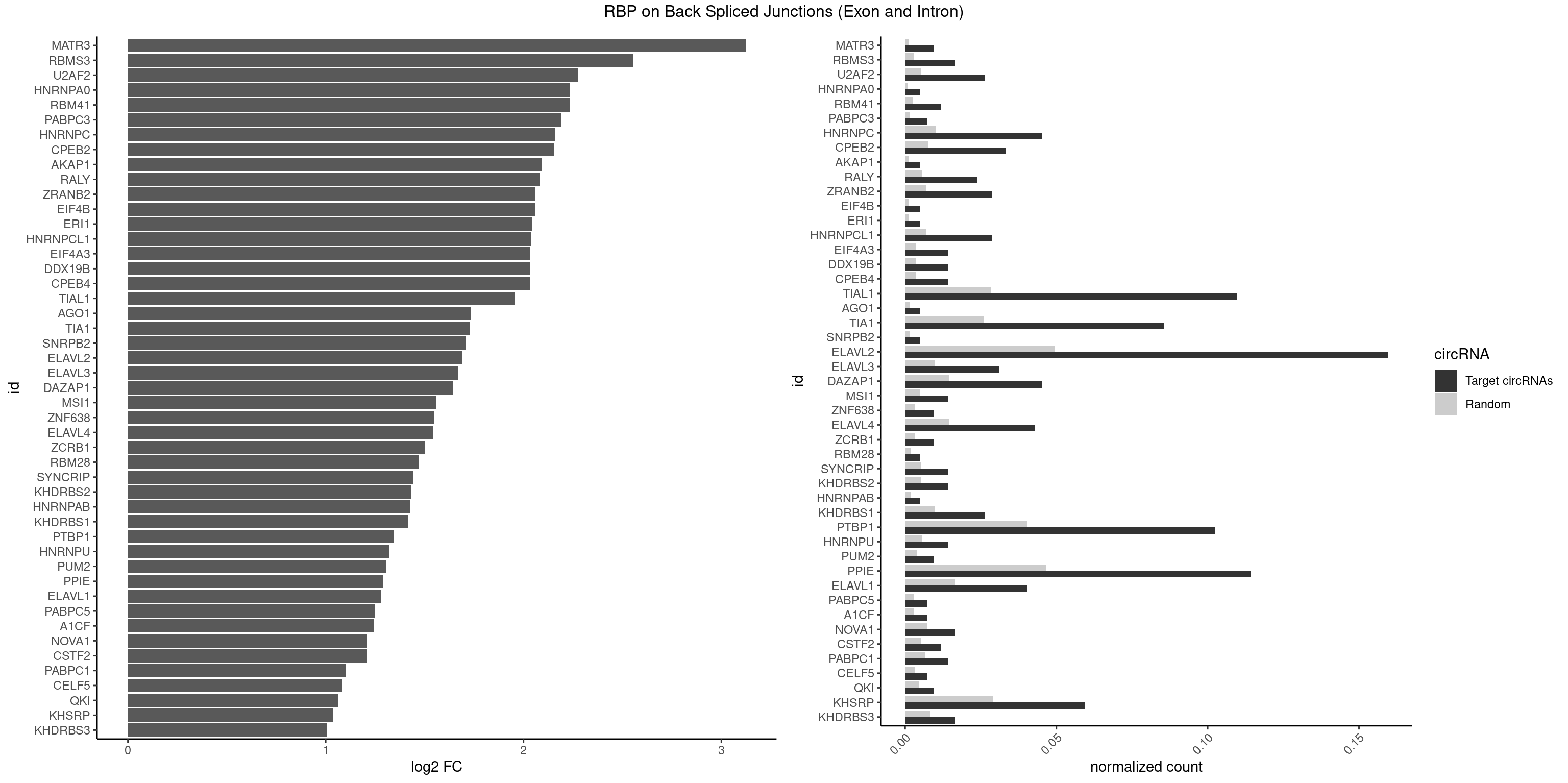
| MATR3 |
3 |
216 |
0.009523810 |
0.001093309 |
3.122837 |
AUCUUA,CAUCUU |
AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMS3 |
6 |
562 |
0.016666667 |
0.002836558 |
2.554752 |
AAUAUA,AUAUAU,CUAUAG,UAUAGC |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| U2AF2 |
10 |
1071 |
0.026190476 |
0.005401048 |
2.277731 |
UUUUCC,UUUUUC,UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| HNRNPA0 |
1 |
200 |
0.004761905 |
0.001012696 |
2.233337 |
AAUUUA |
AAUUUA,AGAUAU,AGUAGG |
| RBM41 |
4 |
502 |
0.011904762 |
0.002534260 |
2.231902 |
UACAUU,UACUUU,UUACUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PABPC3 |
2 |
310 |
0.007142857 |
0.001566909 |
2.188580 |
AAAACA,GAAAAC |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPC |
18 |
2006 |
0.045238095 |
0.010111850 |
2.161491 |
AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CPEB2 |
13 |
1487 |
0.033333333 |
0.007496977 |
2.152585 |
AUUUUU,CAUUUU,CUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| AKAP1 |
1 |
221 |
0.004761905 |
0.001118501 |
2.089973 |
AUAUAU |
AUAUAU,UAUAUA |
| RALY |
9 |
1117 |
0.023809524 |
0.005632809 |
2.079612 |
UUUUUC,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| ZRANB2 |
11 |
1360 |
0.028571429 |
0.006857114 |
2.058900 |
AAAGGU,AGGUAA,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GGUAAA,GUAAAG |
AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| EIF4B |
1 |
226 |
0.004761905 |
0.001143692 |
2.057840 |
UUGGAC |
CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 |
1 |
228 |
0.004761905 |
0.001153769 |
2.045185 |
UUUCAG |
UUCAGA,UUUCAG |
| HNRNPCL1 |
11 |
1381 |
0.028571429 |
0.006962918 |
2.036809 |
AUUUUU,CUUUUU,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB4 |
5 |
691 |
0.014285714 |
0.003486497 |
2.034723 |
UUUUUU |
UUUUUU |
| DDX19B |
5 |
691 |
0.014285714 |
0.003486497 |
2.034723 |
UUUUUU |
UUUUUU |
| EIF4A3 |
5 |
691 |
0.014285714 |
0.003486497 |
2.034723 |
UUUUUU |
UUUUUU |
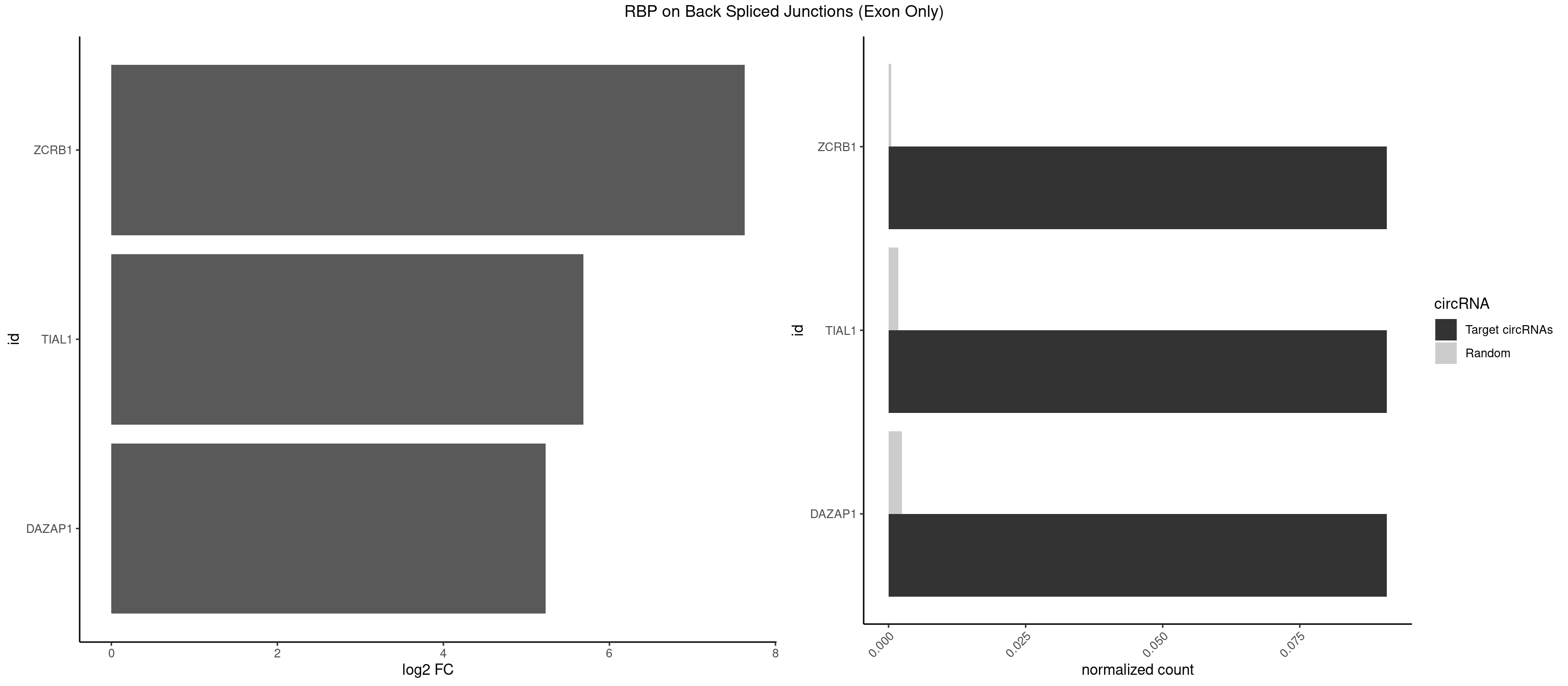
| TIAL1 |
45 |
5597 |
0.109523810 |
0.028204353 |
1.957255 |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CUUUUA,CUUUUU,GUUUUU,UAAAUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| AGO1 |
1 |
283 |
0.004761905 |
0.001430875 |
1.734641 |
AGGUAG |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| TIA1 |
35 |
5140 |
0.085714286 |
0.025901854 |
1.726480 |
AUUUUC,AUUUUG,AUUUUU,CUUUUA,CUUUUU,GUUUUA,GUUUUU,UAUUUU,UUAUUU,UUUUAU,UUUUCU,UUUUGG,UUUUUA,UUUUUC,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SNRPB2 |
1 |
288 |
0.004761905 |
0.001456066 |
1.709463 |
GUAUUG |
AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ELAVL2 |
66 |
9826 |
0.159523810 |
0.049511286 |
1.687942 |
AAUUUA,AAUUUU,AUAUUU,AUUAUU,AUUUAG,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUUCU,CUUUUA,CUUUUU,GAUUUU,GUAUUG,GUUUUA,GUUUUU,UACUUU,UAUAUU,UAUGUU,UAUUAU,UAUUGU,UAUUUU,UGAUUU,UGUAUU,UUAAGU,UUACUU,UUAUUU,UUUAAG,UUUAAU,UUUAUA,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL3 |
12 |
1928 |
0.030952381 |
0.009718863 |
1.671191 |
AUUUUA,UAUUUU,UUAUUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| DAZAP1 |
18 |
2878 |
0.045238095 |
0.014505240 |
1.640964 |
AAUUUA,AGGAAA,AGGUAA,AGGUAG,AGGUUA,AGUUUA,UAGGAA,UAGGUA,UAGGUU,UAGUAU,UAGUUU,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| MSI1 |
5 |
961 |
0.014285714 |
0.004846836 |
1.559458 |
AGGUAG,UAGGAA,UAGGUA,UAGUUG |
AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ZNF638 |
3 |
646 |
0.009523810 |
0.003259774 |
1.546767 |
GUUCUU,GUUGUU,UGUUCU |
CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ELAVL4 |
17 |
2916 |
0.042857143 |
0.014696695 |
1.544044 |
GUAUCU,UAUUUU,UUAUUU,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| ZCRB1 |
3 |
666 |
0.009523810 |
0.003360540 |
1.502846 |
ACUUAA,GACUUA,GUUUAA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM28 |
1 |
340 |
0.004761905 |
0.001718057 |
1.470761 |
AGUAGU |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SYNCRIP |
5 |
1041 |
0.014285714 |
0.005249899 |
1.444212 |
UUUUUU |
AAAAAA,UUUUUU |
| KHDRBS2 |
5 |
1051 |
0.014285714 |
0.005300282 |
1.430432 |
UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPAB |
1 |
351 |
0.004761905 |
0.001773478 |
1.424957 |
GACAAA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| KHDRBS1 |
10 |
1945 |
0.026190476 |
0.009804514 |
1.417524 |
CAAAAU,GAAAAC,UUAAAU,UUUUAC,UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| PTBP1 |
42 |
8003 |
0.102380952 |
0.040326481 |
1.344148 |
ACUUUC,ACUUUU,AUUUUC,AUUUUU,CAUCUU,CUCCAU,CUUUCU,CUUUUU,UACUUU,UAUUUU,UCCUCU,UCUUCU,UUACUU,UUAUUU,UUCUUC,UUCUUG,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| HNRNPU |
5 |
1136 |
0.014285714 |
0.005728537 |
1.318335 |
UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| PUM2 |
3 |
764 |
0.009523810 |
0.003854293 |
1.305073 |
GUACAU,UAAAUA,UGUAUA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PPIE |
47 |
9262 |
0.114285714 |
0.046669690 |
1.292087 |
AAAUAU,AAAUUU,AAUAUA,AAUUAU,AAUUUA,AAUUUU,AUAAUU,AUAUAU,AUAUUU,AUUAUU,AUUUUA,AUUUUU,UAAAUA,UAAAUU,UAAUUA,UAUAUU,UAUUAU,UAUUUU,UUAAAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| ELAVL1 |
16 |
3309 |
0.040476190 |
0.016676743 |
1.279236 |
UAGUUU,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAUUU,UUUGGU,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| PABPC5 |
2 |
596 |
0.007142857 |
0.003007860 |
1.247764 |
AGAAAA,GAAAUU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF |
2 |
598 |
0.007142857 |
0.003017936 |
1.242939 |
AUAAUU,UAAUUA |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| NOVA1 |
6 |
1428 |
0.016666667 |
0.007199718 |
1.210953 |
AUUCAU,UUUUUU |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| CSTF2 |
4 |
1022 |
0.011904762 |
0.005154172 |
1.207726 |
GUGUUU,UGUGUU,UGUUUG,UGUUUU |
GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| PABPC1 |
5 |
1321 |
0.014285714 |
0.006660621 |
1.100845 |
ACGAAU,ACUAAU,AGAAAA,CGAAUC,GAAAAC |
AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| CELF5 |
2 |
669 |
0.007142857 |
0.003375655 |
1.081334 |
GUGUUU,UGUGUU |
GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| QKI |
3 |
904 |
0.009523810 |
0.004559653 |
1.062615 |
ACUAAU,ACUUAU,UCUACU |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| KHSRP |
24 |
5764 |
0.059523810 |
0.029045748 |
1.035140 |
AUAUUU,AUGUAU,AUUAUU,AUUUUA,CUGUGU,GUGUAU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUU,UGUAUA,UGUGUA,UUAUUA,UUAUUU,UUUAUA |
ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| KHDRBS3 |
6 |
1646 |
0.016666667 |
0.008298065 |
1.006119 |
UAAAUA,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |