circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000243156:-:22:17864898:17906886
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000243156:-:22:17864898:17906886 | ENSG00000243156 | ENST00000672019 | - | 22 | 17864899 | 17906886 | 2679 | AUUAACGUGACAGUGUCAGGUGGACCCACAGCCAUCCCACCUCCCCCUCUGGGGAGUGCUGAGAGUGAGGCAGCAUGGAGGAGAGGAAGCAUGAGACCAUGAACCCAGCUCAUGUCCUCUUUGACCGGUUUGUCCAGGCCACCACCUGCAAGGGAACCCUCAAGGCUUUCCAGGAGCUCUGUGACCACCUGGAACUAAAGCCAAAGGACUACCGCUCCUUCUAUCACAAGCUCAAGUCCAAGCUUAACUACUGGAAAGCCAAAGCCCUCUGGGCAAAAUUGGACAAACGGGGCAGUCACAAAGACUACAAAAAGGGAAAAGCGUGCACUAACACCAAGUGUCUCAUCAUUGGGGCUGGCCCCUGUGGUCUCCGUACAGCCAUCGACUUAUCCUUACUGGGGGCCAAGGUGGUUGUUAUUGAGAAACGAGAUGCCUUCUCCCGCAACAACGUCUUGCAUCUCUGGCCAUUCACCAUACAUGAUCUACGAGGUCUGGGUGCCAAGAAGUUCUAUGGCAAGUUCUGUGCUGGAGCCAUCGACCAUAUCAGUAUCCGUCAGCUCCAACUAAUACUUUUGAAAGUAGCCUUGAUCCUAGGCAUUGAAAUCCACGUCAAUGUGGAAUUCCAAGGACUUAUACAGCCUCCUGAGGACCAAGAGAAUGAACGGAUAGGCUGGCGGGCACUGGUGCACCCCAAGACUCAUCCUGUGUCAGAGUAUGAAUUUGAAGUGAUCAUCGGUGGGGAUGGUCGGAGGAACACCUUGGAAGGGUUUCGUCGGAAAGAAUUCCGUGGCAAACUGGCCAUCGCCAUCACGGCAAAUUUUAUCAACCGAAAUACAACAGCAGAAGCUAAAGUGGAAGAGAUCAGUGGUGUGGCUUUUAUAUUCAACCAAAAAUUUUUCCAGGAACUGAGGGAAGCCACAGGUAUUGACUUGGAGAACAUCGUUUACUACAAAGAUGACACACACUAUUUCGUUAUGACAGCCAAAAAGCAGAGUUUGCUGGACAAAGGAGUGAUACUACAUGACUACGCCGACACAGAGCUCCUGCUUUCCCGAGAAAACGUGGACCAGGAGGCUCUGCUCAGCUAUGCCAGGGAGGCGGCAGACUUCUCUACCCAGCAGCAGCUGCCGUCUCUGGAUUUUGCCAUCAAUCACUAUGGGCAGCCCGAUGUGGCCAUGUUUGACUUCACUUGUAUGUAUGCCUCCGAGAACGCCGCCUUGGUGCGGGAGCAGAACGGACACCAGUUACUAGUGGCUCUGGUCGGGGACAGCCUCCUAGAGCCUUUCUGGCCAAUGGGAACAGGAAUAGCCCGGGGCUUUCUAGCUGCUAUGGACUCUGCCUGGAUGGUCCGAAGUUGGUCUCUAGGAACGAGCCCUUUGGAAGUGCUGGCAGAGAGGGAAAGUAUUUACAGGUUGCUGCCUCAGACCACCCCUGAGAAUGUGAGUAAGAACUUCAGCCAGUACAGUAUCGACCCUGUCACUCGGUAUCCCAAUAUCAACGUCAACUUCCUCCGGCCAAGCCAGGUGCGCCAUUUAUAUGAUACUGGCGAAACAAAAGAUAUUCACCUGGAAAUGGAGAGCCUGGUGAAUUCCCGAACCACCCCCAAAUUGACUCGCAAUGAGUCUGUAGCUCGUUCAAGCAAACUGCUGGGUUGGUGCCAGAGGCAGACAGAUGGCUAUGCAGGGGUAAACGUGACAGAUCUCACCAUGUCCUGGAAAAGUGGCUUGGCCCUUUGUGCAAUUAUCCAUAGAUACCGCCCUGACCUGAUAGAUUUUGAUUCUUUGGAUGAGCAAAAUGUGGAGAAGAAUAACCAACUGGCCUUUGACAUUGCUGAGAAGGAAUUGGGCAUUUCUCCCAUCAUGACAGGCAAAGAAAUGGCCUCCGUGGGGGAGCCUGAUAAGCUGUCCAUGGUGAUGUACCUGACUCAGUUCUACGAGAUGUUUAAGGACUCCCUCCCCUCUAGCGACACCUUGGACCUAAAUGCCGAGGAGAAAGCAGUCCUGAUAGCCAGCACCAGAUCCCCUAUCUCCUUCCUAAGCAAACUUGGCCAGACCAUCUCUCGGAAGCGUUCUCCCAAGGAUAAAAAGGAAAAGGACUUGGAUGGUGCUGGGAAGAGGAGAAAGACCAGUCAAUCAGAGGAGGAGGAAGCUCCUCGGGGCCACAGAGGAGAAAGACCGACCCUGGUGAGCACUCUGACAGACAGGAGGAUGGACGUUGCCGUUGGGAACCAGAACAAAGUGAAGUACAUGGCGACCCAGCUGCUGGCCAAAUUUGAAGAGAAUGCGCCCGCACAGUCCAUCGGCAUACGGAGACAGGGCUCCAUGAAGAAGGAGUUCCCGCAGAACCUGGGAGGCAGCGACACAUGCUACUUCUGCCAGAAGCGGGUCUACGUGAUGGAGAGGCUGAGUGCCGAGGGCAAGUUCUUCCACCGGAGCUGCUUCAAGUGCGAGUACUGCGCCACCACCCUGCGCCUCUCGGCCUACGCCUACGACAUCGAGGAUGGUAAAUUCUACUGUAAGCCACACUACUGCUAUCGACUCUCUGGCUACGCACAAAGGAAGAGACCGGCAGUGGCUCCCCUGUCUGGAAAGGAGGCCAAAGGACCCCUGCAGGAUGGCGCCACCACAGAUGCAAACGGACGGGCCAACGCCGUGGCCAGCUCCACUGAGAGAACCCCAGAUUAACGUGACAGUGUCAGGUGGACCCACAGCCAUCCCACCUCCCCCUCU | circ |
| ENSG00000243156:-:22:17864898:17906886 | ENSG00000243156 | ENST00000672019 | - | 22 | 17864899 | 17906886 | 22 | GAGAACCCCAGAUUAACGUGAC | bsj |
| ENSG00000243156:-:22:17864898:17906886 | ENSG00000243156 | ENST00000672019 | - | 22 | 17906877 | 17907086 | 210 | GAUCCUUGAGAGCAAGAACCAUCCUGCUCACCUUUCCAGCCCCAGUGUUUCGUAGUGCCUGGAGUAUUGCAGGUGUGCAGUCCACACUUAUGGAUGGAUGACUGCGACACUUUAGAACUCUGCUUUGAAGAGGAAUAUGUCUCCUGGAGAAUGUUUGUAGACAGAGAAAUGCUAACCACGUUUUUCUUUCUUUUUGUCAGAUUAACGUGA | ie_up |
| ENSG00000243156:-:22:17864898:17906886 | ENSG00000243156 | ENST00000672019 | - | 22 | 17864699 | 17864908 | 210 | AGAACCCCAGGUAGCCUCACUUCCUUGUUUGGCUGGGUGGCGCGUCACUCCCUCGGCCUCUGUGACAAGGCCAAGGGCAUGAGCCAGCACCUCCAAAGCAACAUUUCUUCAUUCGGGCAACAAGUGGCCCAAAACCCUCUGGACUCCUUUUUCAUGUGCCAGCUGCUUGCGUUCGGAGUCCCCUUUCUCUAUGGCCUGUCGGAGGUGCUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
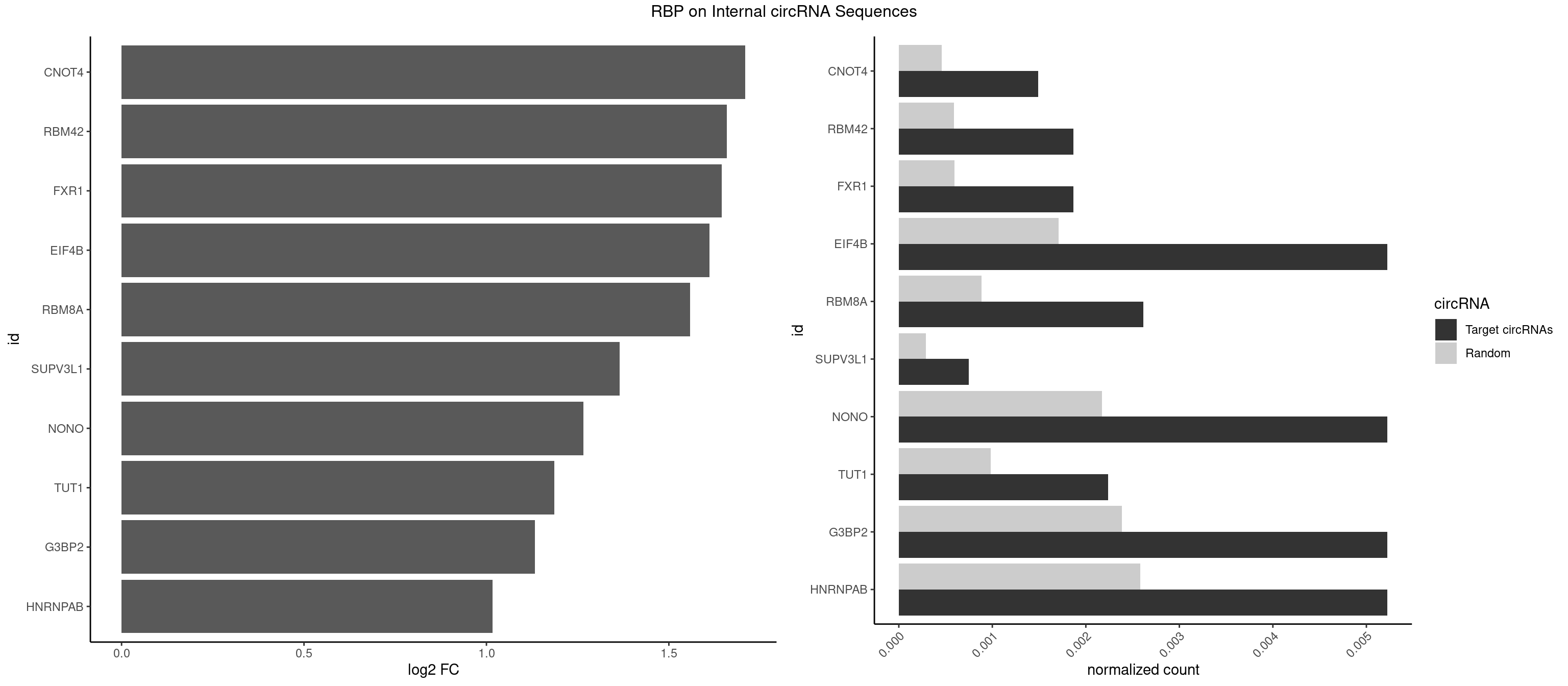
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 3 | 314 | 0.0014930944 | 0.0004564992 | 1.709621 | GACAGA | GACAGA |
| RBM42 | 4 | 407 | 0.0018663680 | 0.0005912752 | 1.658332 | AACUAA,AACUAC,ACUACG | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR1 | 4 | 411 | 0.0018663680 | 0.0005970720 | 1.644257 | ACGACA,AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| EIF4B | 13 | 1179 | 0.0052258305 | 0.0017100607 | 1.611613 | CUCGGA,CUUGGA,GUCGGA,UCGGAA,UUGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM8A | 6 | 611 | 0.0026129153 | 0.0008869128 | 1.558796 | AUGCGC,GUGCGC,UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| SUPV3L1 | 1 | 199 | 0.0007465472 | 0.0002898408 | 1.364973 | CCGCCC | CCGCCC |
| NONO | 13 | 1498 | 0.0052258305 | 0.0021723567 | 1.266399 | AGAGGA,AGGAAC,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| TUT1 | 5 | 678 | 0.0022396417 | 0.0009840095 | 1.186524 | AAAUAC,AAUACU,AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| G3BP2 | 13 | 1644 | 0.0052258305 | 0.0023839405 | 1.132312 | AGGAUA,AGGAUG,GGAUAA,GGAUAG,GGAUGA,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPAB | 13 | 1782 | 0.0052258305 | 0.0025839306 | 1.016093 | AAAGAC,ACAAAG,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
RBP on BSJ (Exon Only)
Plot
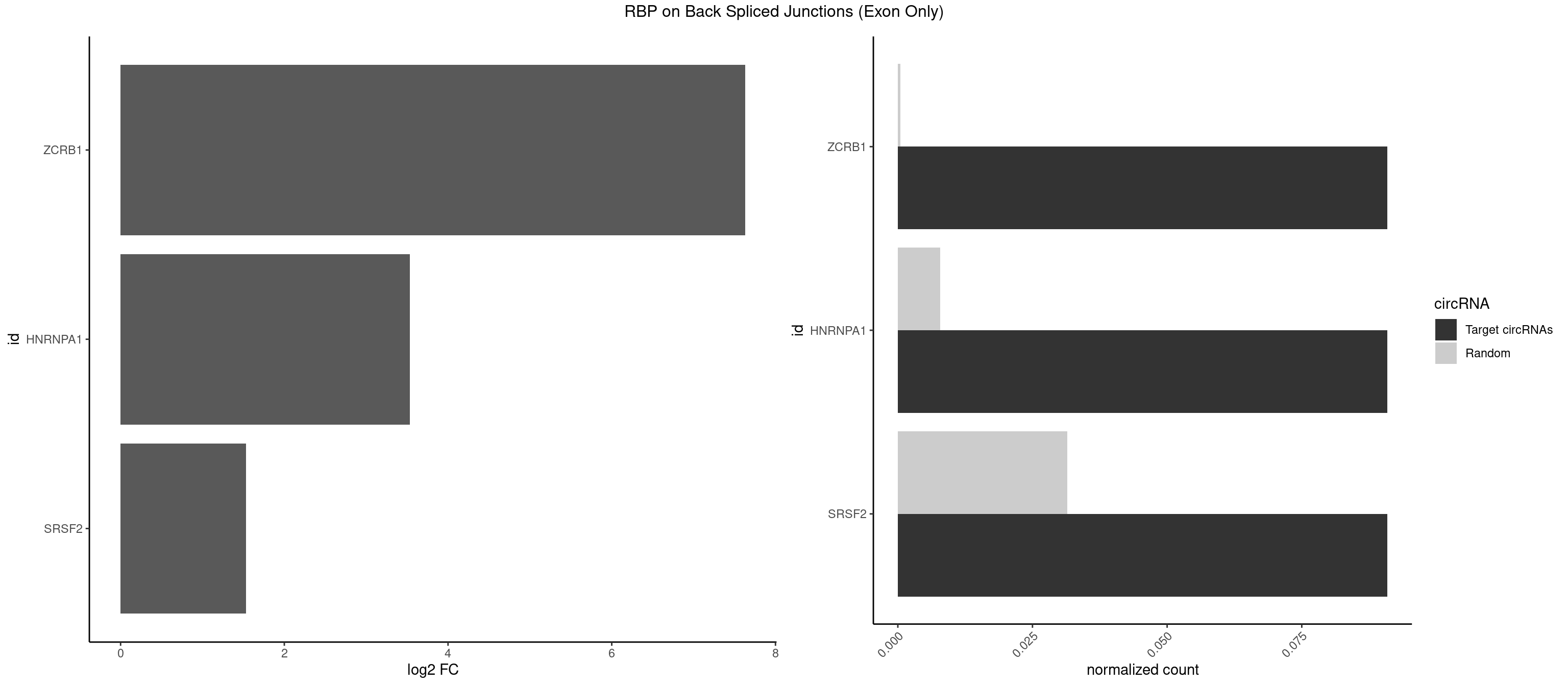
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZCRB1 | 1 | 6 | 0.09090909 | 0.0004584752 | 7.631437 | GAUUAA | AAUUAA,AUUUAA,GACUUA,GGCUUA |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.0078595756 | 3.531901 | AGAUUA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
| SRSF2 | 1 | 479 | 0.09090909 | 0.0314383023 | 1.531901 | CCAGAU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
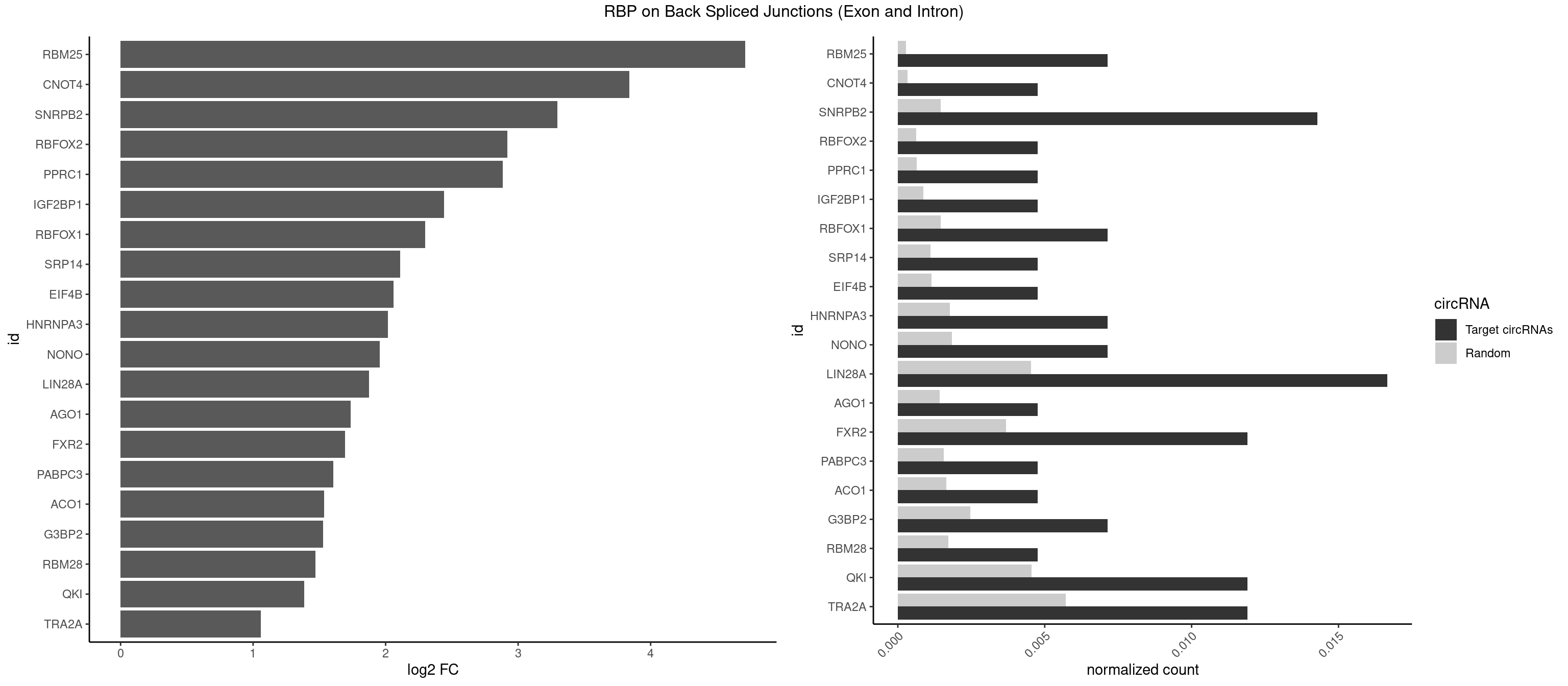
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 2 | 53 | 0.007142857 | 0.0002720677 | 4.714464 | CGGGCA,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| SNRPB2 | 5 | 288 | 0.014285714 | 0.0014560661 | 3.294425 | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| PPRC1 | 1 | 127 | 0.004761905 | 0.0006449012 | 2.884389 | GGCGCG | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGA,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | GUCGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| NONO | 2 | 364 | 0.007142857 | 0.0018389762 | 1.957598 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| LIN28A | 6 | 900 | 0.016666667 | 0.0045395002 | 1.876360 | CGGAGG,CGGAGU,GGAGAA,GGAGUA,UGGAGA,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | AGGUAG | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| FXR2 | 4 | 730 | 0.011904762 | 0.0036829907 | 1.692589 | AGACAG,GACAAG,GACAGA,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | GGAUGA,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACUUAU,AUUAAC,CACACU,CUAACC | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AAGAGG,AGAGGA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.