circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000288637:+:4:153322647:153394336
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000288637:+:4:153322647:153394336 | ENSG00000288637 | ENST00000674680 | + | 4 | 153322648 | 153394336 | 2351 | ACAAAAAUUGGAUCAGGAAAGCUGAUGGGACCCAAAGGAGUUUCUGUGGACCGCAAUGGGCACAUUAUUGUUGUGGACAACAAGGCGUGCUGCGUGUUUAUCUUCCAGCCAAACGGGAAAAUAGUCACCAGGUUUGGUAGCCGAGGAAAUGGGGACAGGCAGUUUGCAGGUCCCCAUUUUGCAGCUGUAAAUAGCAAUAAUGAGAUUAUUAUUACAGAUUUCCAUAAUCAUUCUGUCAAGGUGUUUAAUCAGGAAGGAGAAUUCAUGUUGAAGUUUGGCUCAAAUGGAGAAGGAAAUGGGCAGUUUAAUGCUCCAACAGGUGUAGCAGUGGAUUCAAAUGGAAACAUCAUUGUGGCCGACUGGGGAAACAGCAGGAUCCAGCUCCUCCUGCCAGGCACGGUGGCUCACGCCUGUAAUCCUAGCACUUUAGGAGGCCAAGGCGGAUGGGUCGCCUGAGACUGGGAGAUCGAGACCAGCCUGGCCAACAUGGUGGCGGUGAUUCCCAGGUCUGGUUGAUCAAGACUGCAAUGCACAACAGGAACCUCAGAGAACUGUCACAGCAGCUCUCAGUGACACUUAGGAUACAGAUGCAGCUUAAGUGACCCUUUUUUAUAUCCAUUUAGCACUCAUUUUUCCCAAAAGAAGACCUGUUUGGGUCAAUUUGUUACUGUCCCAGGGCAGGCUUUUGCACUGUGUCGCUUUAGAGGCAGGGGCGGAGGAAAACUUGGGACUGUAACAUCAGUACUGCACAAAGCCCUCUUCCGUAUACAGCAUAAGCACUAUUCUAUAAAAUCCCCAGCAAGCCUUUGUUUCCUUGCAGUCAGCUCCUUUUCUGCUGAUUCUGCCCAUUGCUCCCUUGCAACGUAUUUUCAUGCUUUCUCUAAUGAUUCUGCCUUUCUUUACUUACAACUGCCUUGGUAAAUUUUUCUGCCACAGCGCCACUGGCCUCAGAUAGUCACCACUCACCUGCAACAAGCCUGAGAGCCAAUUAAGACAAGGCUUUCCCCAAGUAGCACAAAUGUAUAUACCACUACUGUAGUUACUUAAUGUACUCAUUUAUCAUUUUAAAAUUACCUUAUGUACCAUCAGUUACAUGGUAUAACUCUUACAGGAAAAACCAAAUUAAUGGAACUCUGCAAUUCAAUGUACCCCUUCCUCUGCCUCCAGCAGAAGCAACAGGGGACAUAAAUUCCAACUUUAGAUGCUACUCUUAUAGCAUCUACAAGACUUACUACACUGUGUGUGUGUGUGUGUGUGUGUGUGCGUGCGUGUGUGUGUGUUACAGAGUCUCACUCUGUCGCCCAGGCUGGAGUGCAGUGGCGGGGUCUUGGCUCACUGCAACCUCUGCCUCCUGGGUCCAAGCGAUUCUCCUGCCUCAGCCUCCCGAGUAGCUGGGAUUGUACAGGCGUCCGCCACCACGCCUGGCUAAUUUUUGUAUUUUUAUUAGAGAUGGGAUUUCACCAUGUUGGCCAGGCUGGUCUCAAACUCCCUACCUCAAGUGAUUUGCCCGCCUCGGCCUCCCAAAUUGCUGGGAUUACAGGUGUGAGCCACAGCGCCCAACCGUACUUCUUACACCUAAAAAUGGUUCAAAACACCAUAGAAUAAUUUUCAAAAUAAUUGAAUUCUGAGUUAAAAUUUUUCAGCACUUUCAGGGGCCUGAGCUUUUGCUAUGAAUCUGGAUCUAUUCUCAAGCCAUGAAGUAGCGUAUUUUUGACCAUGUUGAUUUGAAAUUCAUUCCGUUGUUUCCUUCUUUUUUAAAACCUAAGGGAAAAACAAACAGACAUACACAGGAUUUGAAAAGCUGGAAAGCAGUGGGUAGCGGCUCGGGAGAAGAUGAGGGGUCAGAUUAGAAGACGAGAAAGGAAUCCAGUCCGCCCCAGACAGGACAAGUCCGGAAGGAUGGAGGCAGCGUUUCCUUCCUCAACAGCCUAAACGCCUCUAUUUUUUACAUCCAAACUUAGGCCAUAUUUCUGCGGCGCCAAGAAAACGCAGUAAAAGCAAAGUCAAAGAAAAAAGGACUGAGUGCAGAAGAAAAGAGAACUCGCAUGAUGGAAAUAUUUUCUGAAACAAAAGAUGUAUUUCAAUUAAAAGACUUGGAGAAGAUUGCUCCCAAAGAGAAAGGCAUUACUGCUAUGUCAGUAAAAGAAGUCCUUCAAAGCUUAGUUGAUGAUGGUAUGGUUGACUGUGAGAGGAUCGGAACUUCUAAUUAUUAUUGGGCUUUUCCAAGUAAAGCUCUUCAUGCAAGGAAACAUAAGUUGGAGGUUCUGGAAUCUCAGUUGUCUGAGGGAAGUCAAAAGCAUGCAAGCCUACAGAAAAGCAUUGAGAAAGCUAAAAUUGGCCGAUGUGAAACGACAAAAAUUGGAUCAGGAAAGCUGAUGGGACCCAAAGGAGUUUCUGUGGA | circ |
| ENSG00000288637:+:4:153322647:153394336 | ENSG00000288637 | ENST00000674680 | + | 4 | 153322648 | 153394336 | 22 | GAUGUGAAACGACAAAAAUUGG | bsj |
| ENSG00000288637:+:4:153322647:153394336 | ENSG00000288637 | ENST00000674680 | + | 4 | 153322448 | 153322657 | 210 | UAUAUAGAUAUCAGCAGCAGAUAAUGGAUGUGACAAAGCCAGUUUUCCAAGACAGAAGUUCAUUGUGAUAUAUGAGUAGCUGUGUACCCGUUUGAACCUUCAGGUAAAUGUUGACCAAAACAGUGUUUAUCUUCAUGUUCUGUAGUGGUUUAAACUUUGACUCUAUACUGUACUUCUAUUUCUGUACUUGUUUCAAACAGACAAAAAUUG | ie_up |
| ENSG00000288637:+:4:153322647:153394336 | ENSG00000288637 | ENST00000674680 | + | 4 | 153394327 | 153394536 | 210 | AUGUGAAACGGUAAGUUUGUGUCUAUAGAUUAUUUUAAUAAUGGCAAUUCUAAAUAUUAGAGUAAGCGACACAGAGCUUCCUGUGAGGAGGAUGCAUUUCUACCAGAAGAGAAGGGGAUCAUAGCCAAGGAUAGGAACGUUUGACCUGGUAGGAUUCACCAAUUGUGAGUCUAGAUUUUAUUAUUAUUAUAUAUGGCAGCAUAGCAUAGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
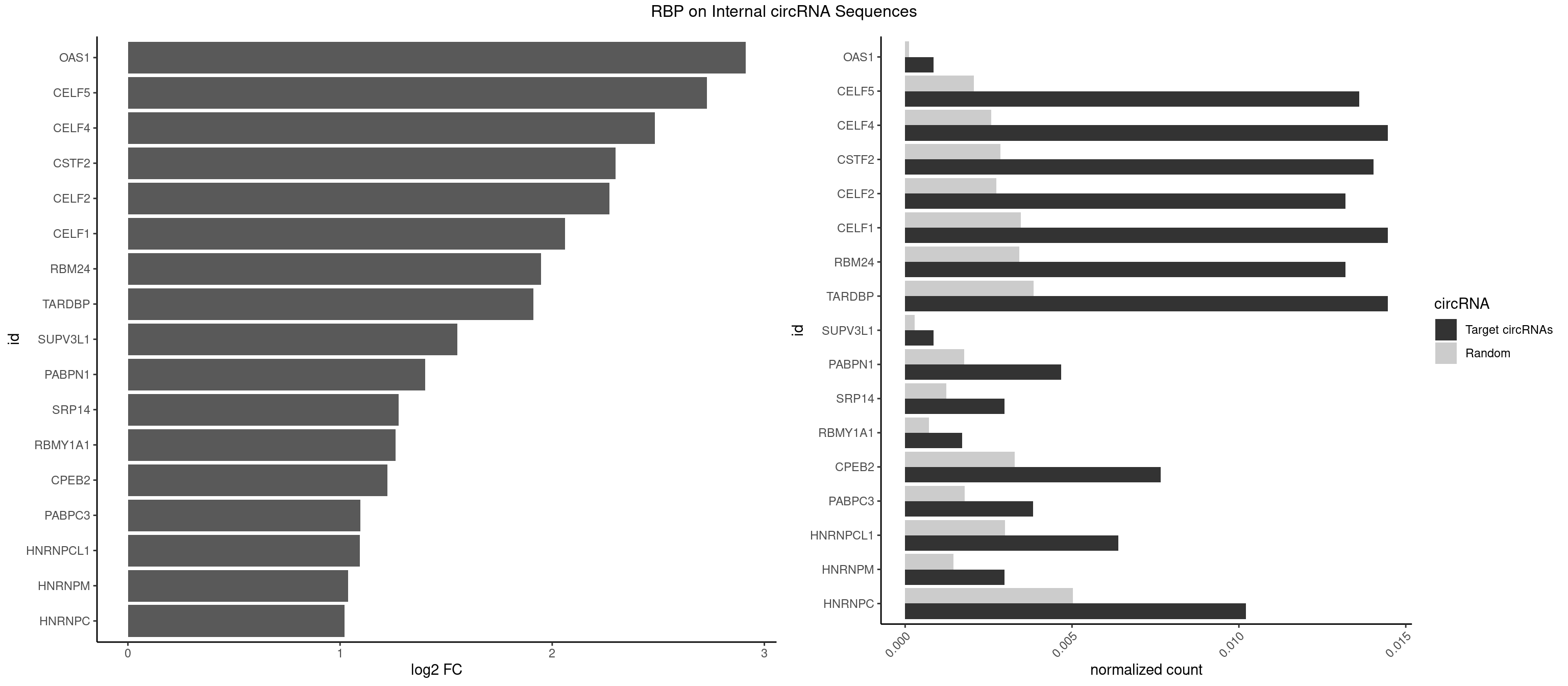
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.0008507018 | 0.0001130379 | 2.911847 | GCAUAA | GCAUAA |
| CELF5 | 31 | 1415 | 0.0136112293 | 0.0020520728 | 2.729644 | GUGUGU,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 33 | 1782 | 0.0144619311 | 0.0025839306 | 2.484621 | GGUGUG,GGUGUU,GUGUGU,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CSTF2 | 32 | 1967 | 0.0140365802 | 0.0028520334 | 2.299129 | GUGUGU,GUGUUU,UGUGUG,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF2 | 30 | 1886 | 0.0131858783 | 0.0027346479 | 2.269567 | GUGUGU,GUUGUU,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF1 | 33 | 2391 | 0.0144619311 | 0.0034664959 | 2.060710 | GUGUGU,GUUGUG,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| RBM24 | 30 | 2357 | 0.0131858783 | 0.0034172229 | 1.948097 | GUGUGA,GUGUGU,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| TARDBP | 33 | 2654 | 0.0144619311 | 0.0038476365 | 1.910216 | GUGUGU,GUUGUG,GUUGUU,UGUGUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SUPV3L1 | 1 | 199 | 0.0008507018 | 0.0002898408 | 1.553393 | CCGCCC | CCGCCC |
| PABPN1 | 10 | 1222 | 0.0046788601 | 0.0017723764 | 1.400472 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| SRP14 | 6 | 847 | 0.0029774564 | 0.0012289250 | 1.276684 | CCUGUA,CGCCUG,CUGUAG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBMY1A1 | 3 | 489 | 0.0017014037 | 0.0007101099 | 1.260611 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| CPEB2 | 17 | 2261 | 0.0076563165 | 0.0032780993 | 1.223791 | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| PABPC3 | 8 | 1234 | 0.0038281582 | 0.0017897669 | 1.096879 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPCL1 | 14 | 2062 | 0.0063802637 | 0.0029897078 | 1.093612 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPM | 6 | 999 | 0.0029774564 | 0.0014492040 | 1.038820 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| HNRNPC | 23 | 3471 | 0.0102084219 | 0.0050316361 | 1.020660 | AUUUUU,CUUUUU,GGAUAC,GGAUUC,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
RBP on BSJ (Exon Only)
Plot
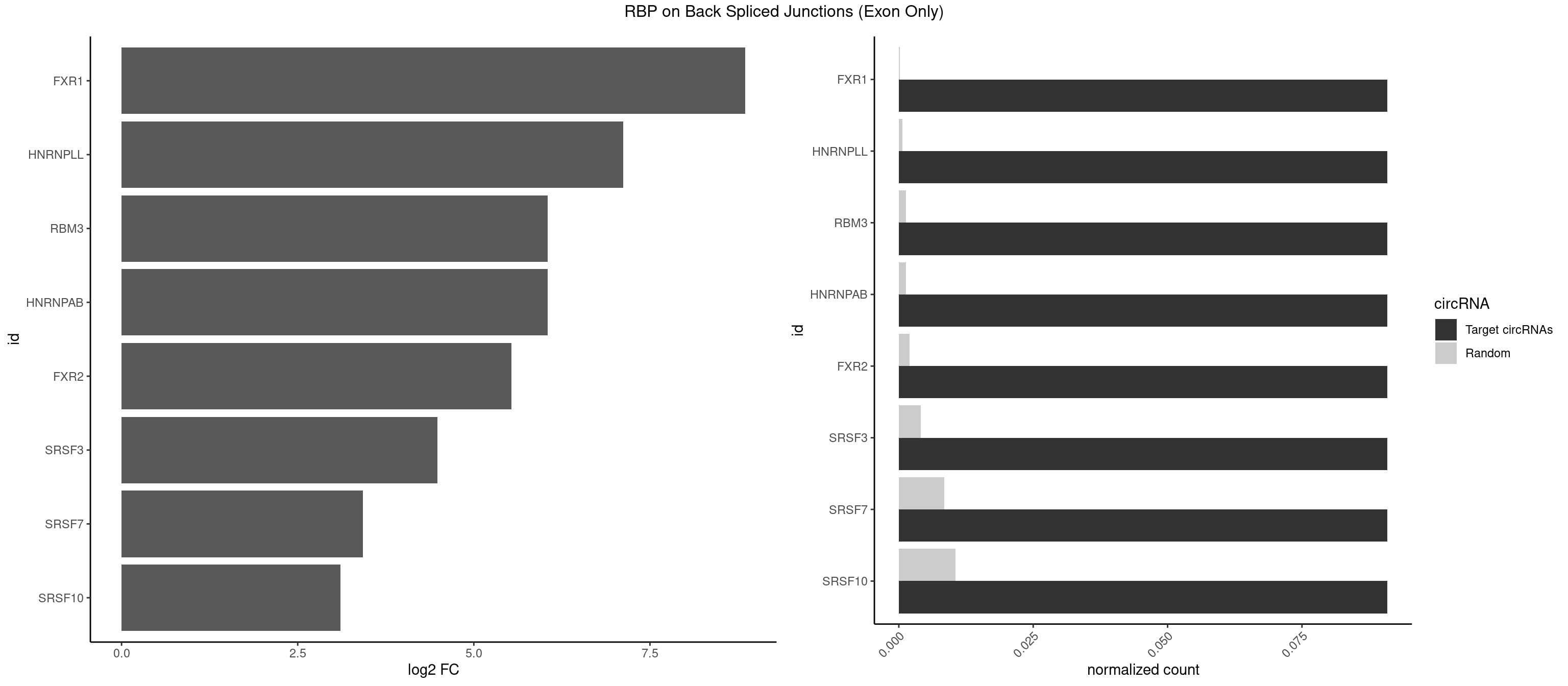
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 2 | 0.09090909 | 0.0001964894 | 8.853829 | ACGACA | ACGACA,AUGACA |
| HNRNPLL | 1 | 9 | 0.09090909 | 0.0006549646 | 7.116864 | ACGACA | ACAAAC,ACACCA,ACAUAC,ACGACA,CACCAC,CAGACG,CAUACA,GACGAC |
| HNRNPAB | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,CAAAGA,GACAAA |
| RBM3 | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | AAACGA | AAAACG,AAAACU,AAGACU,AAUACG,AAUACU,AUACUA,GAAACU,GAGACG,GAGACU,GAUACU |
| FXR2 | 1 | 29 | 0.09090909 | 0.0019648939 | 5.531901 | GACAAA | AGACAA,AGACAG,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGGA,GGACAA,GGACAG,GGACGA,UGACAA,UGACAG,UGACGA |
| SRSF3 | 1 | 61 | 0.09090909 | 0.0040607807 | 4.484596 | AACGAC | AACGAU,ACCACC,ACUACG,AGAGAU,CACAAC,CACAUC,CACCAC,CAGAGA,CAUCAC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CUACAG,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUUAU,GACUUC,GAGAUU,UACAGC,UCAGAG,UCUCCA,UCUUCA,UCUUCC,UGUCAA,UUCGAC,UUCUCC,UUCUUC |
| SRSF7 | 1 | 128 | 0.09090909 | 0.0084490438 | 3.427565 | GACAAA | AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACUACG,AGAAGA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CGAAUG,CUCUUC,CUGAGA,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UAGAGA,UCUUCA,UGAGAG,UGGACA |
| SRSF10 | 1 | 160 | 0.09090909 | 0.0105449306 | 3.107875 | GACAAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon and Intron)
Plot
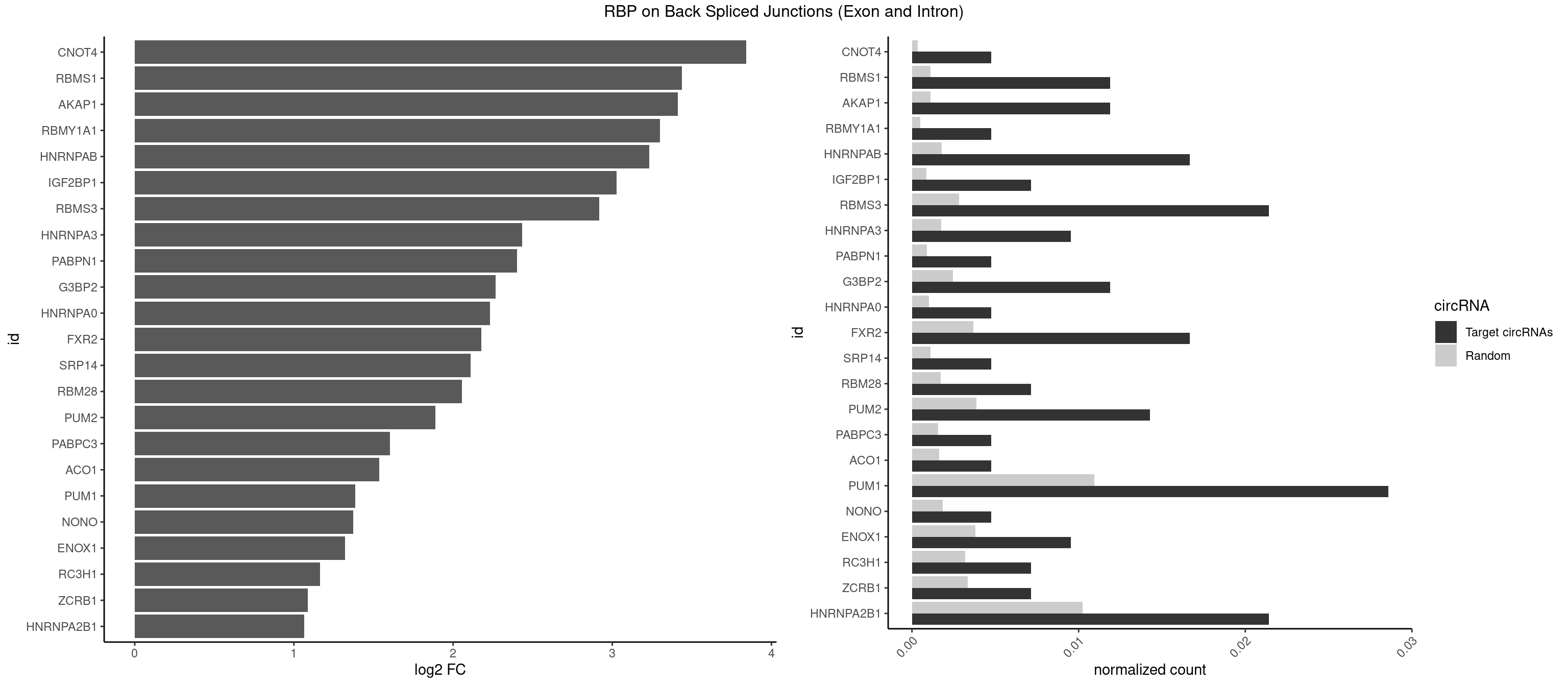
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| RBMS1 | 4 | 217 | 0.011904762 | 0.0010983474 | 3.438132 | AUAUAG,GAUAUA,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 4 | 221 | 0.011904762 | 0.0011185006 | 3.411901 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| HNRNPAB | 6 | 351 | 0.016666667 | 0.0017734784 | 3.232312 | AAGACA,ACAAAG,AGACAA,AUAGCA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| IGF2BP1 | 2 | 173 | 0.007142857 | 0.0008766626 | 3.026408 | ACCCGU,CCCGUU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| RBMS3 | 8 | 562 | 0.021428571 | 0.0028365578 | 2.917322 | AUAUAG,AUAUAU,CUAUAG,UAUAGA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPA3 | 3 | 349 | 0.009523810 | 0.0017634019 | 2.433177 | CAAGGA,CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| G3BP2 | 4 | 490 | 0.011904762 | 0.0024738009 | 2.266737 | AGGAUA,AGGAUG,AGGAUU,GGAUAG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| FXR2 | 6 | 730 | 0.016666667 | 0.0036829907 | 2.178016 | AGACAA,AGACAG,GACAAA,GACAGA,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | GAGUAG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PUM2 | 5 | 764 | 0.014285714 | 0.0038542926 | 1.890035 | GUAAAU,UAAAUA,UAGAUA,UAUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| PUM1 | 11 | 2172 | 0.028571429 | 0.0109482064 | 1.383879 | AAUAUU,AAUGUU,AAUUGU,AGAUAA,CCAGAA,GUAAAU,UAAAUA,UAGAUA,UAUAUA,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AAGACA,AGACAG,CAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPA2B1 | 8 | 2033 | 0.021428571 | 0.0102478839 | 1.064210 | AAGGGG,AGAUAU,AGGAAC,AUAGCA,CAAGGA,CCAAGA,CCAAGG,GCCAAG | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.