circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000070961:-:12:89630504:89642355
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000070961:-:12:89630504:89642355 | ENSG00000070961 | MSTRG.7745.3 | - | 12 | 89630505 | 89642355 | 720 | GUUUAAGUGGAAACCCUGCAGAUUUAGAAAGAAGAGAAGCAGUGUUUGGAAAGAAUUUUAUACCUCCUAAAAAGCCAAAAACCUUUCUUCAAUUAGUAUGGGAAGCAUUACAAGAUGUCACUUUAAUUAUAUUAGAAAUUGCAGCCAUAGUAUCAUUGGGCCUUUCUUUUUAUCAGCCUCCAGAAGGGGAUAAUGCACUUUGUGGAGAAGUUUCUGUUGGGGAGGAAGAAGGUGAAGGUGAAACUGGUUGGAUUGAAGGAGCUGCAAUCCUCUUGUCUGUAGUGUGUGUGGUGUUAGUAACAGCUUUCAAUGACUGGAGUAAGGAAAAACAGUUUAGAGGUUUGCAGAGCCGAAUUGAACAAGAACAGAAGUUCACUGUCAUCAGGGGUGGUCAGGUCAUUCAGAUACCUGUAGCUGACAUUACUGUUGGAGAUAUUGCUCAAGUGAAAUAUGGUGAUCUUCUUCCAGCUGACGGCAUACUUAUUCAAGGCAACGAUCUUAAAAUUGAUGAAAGCUCAUUGACUGGUGAAUCAGAUCAUGUUAAAAAGUCUUUAGAUAAGGAUCCCUUACUUCUAUCAGGUACUCAUGUAAUGGAAGGCUCUGGAAGAAUGGUAGUUACAGCUGUAGGUGUAAAUUCUCAAACUGGAAUUAUCUUUACCUUACUUGGAGCUGGAGGUGAAGAGGAAGAGAAGAAAGAUGAGAAGAAAAAGGAAAAGAAAAGUUUAAGUGGAAACCCUGCAGAUUUAGAAAGAAGAGAAGCAGUGUUUGGA | circ |
| ENSG00000070961:-:12:89630504:89642355 | ENSG00000070961 | MSTRG.7745.3 | - | 12 | 89630505 | 89642355 | 22 | GGAAAAGAAAAGUUUAAGUGGA | bsj |
| ENSG00000070961:-:12:89630504:89642355 | ENSG00000070961 | MSTRG.7745.3 | - | 12 | 89642346 | 89642555 | 210 | CAGUCCUAACUGAUCUACUUAAAGUGUUCUUUUAAAGACCUGCACAUGUUUUUCAGUGAUUUAAAUUCUGUACACUUAAAGCAUGAAAGUAAACAUUUUGGUAGGGUCUAGAGUGAUGAGUUGAGGUAUUUUGAAUAAAACUAUAUAUUUUCAUUUGUAAGAAGUAAAACUGUUCACUGAAAUUUGUUUCUUUUUAUUAGGUUUAAGUGG | ie_up |
| ENSG00000070961:-:12:89630504:89642355 | ENSG00000070961 | MSTRG.7745.3 | - | 12 | 89630305 | 89630514 | 210 | GAAAAGAAAAGUAAGAAUAACAAUUUUCUAUAAUUGGUAUUUGGAAAUAGGGCAAAUAAUUUGUAAGAACUAGUACCUUCUUUCAAGAGAUUCUGGUAUGAAAAAUUCUACAAAAAGUUAAAAGAAGAUUUUUCUAUUAGACUUAAUAGCAUGGGCCUUUAGAGAAUACAUCAGUCCCCUUUACCAUGGCCAGAGAAAUUCAGCAUCUAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
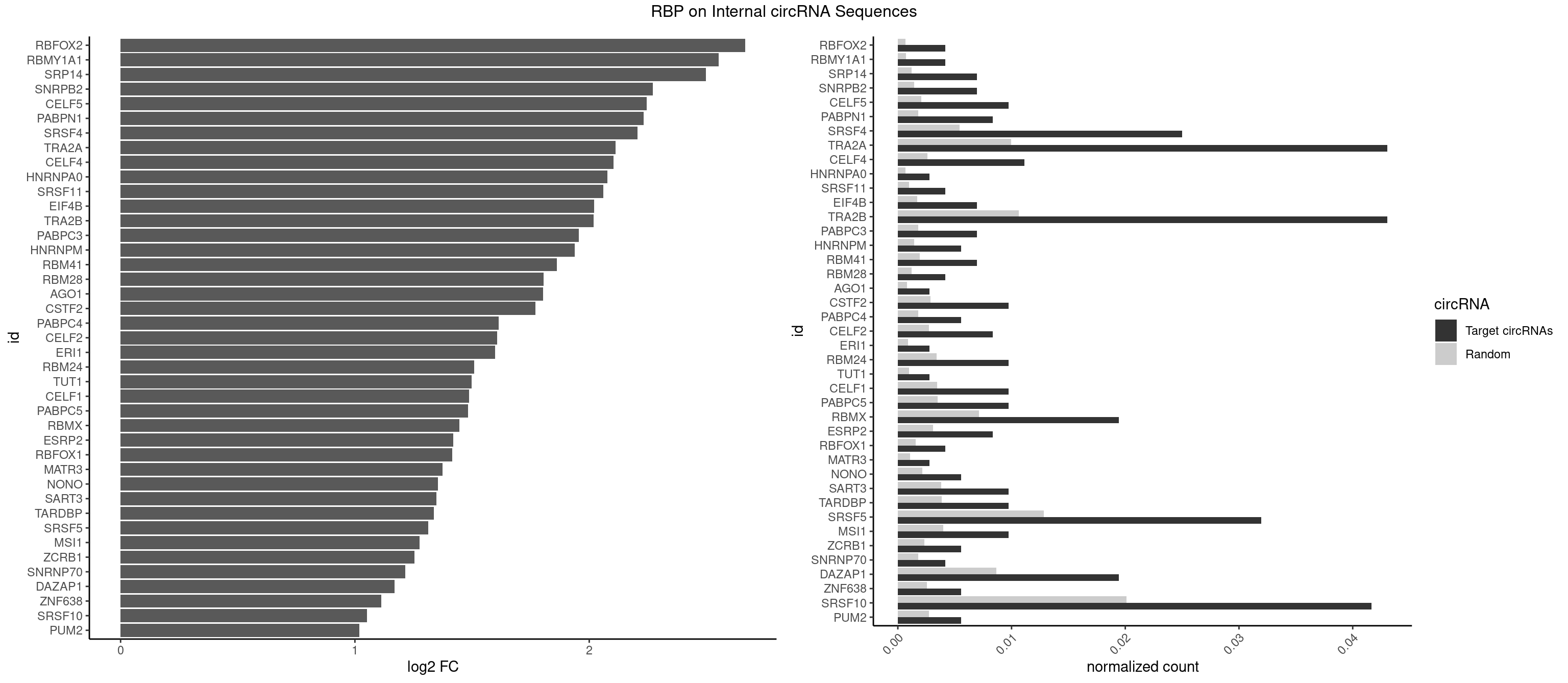
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 2 | 452 | 0.004166667 | 0.0006564894 | 2.666050 | UGACUG | UGACUG,UGCAUG |
| RBMY1A1 | 2 | 489 | 0.004166667 | 0.0007101099 | 2.552779 | ACAAGA | ACAAGA,CAAGAC |
| SRP14 | 4 | 847 | 0.006944444 | 0.0012289250 | 2.498462 | CCUGUA,CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| SNRPB2 | 4 | 991 | 0.006944444 | 0.0014376103 | 2.272187 | AUUGCA,UAUUGC,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| CELF5 | 6 | 1415 | 0.009722222 | 0.0020520728 | 2.244204 | GUGUGG,GUGUGU,GUGUUU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPN1 | 5 | 1222 | 0.008333333 | 0.0017723764 | 2.233209 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| SRSF4 | 17 | 3740 | 0.025000000 | 0.0054214720 | 2.205172 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| TRA2A | 30 | 6871 | 0.043055556 | 0.0099589296 | 2.112137 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| CELF4 | 7 | 1782 | 0.011111111 | 0.0025839306 | 2.104364 | GGUGUU,GUGUGG,GUGUGU,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPA0 | 1 | 453 | 0.002777778 | 0.0006579386 | 2.077906 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| SRSF11 | 2 | 688 | 0.004166667 | 0.0009985015 | 2.061057 | AAGAAG | AAGAAG |
| EIF4B | 4 | 1179 | 0.006944444 | 0.0017100607 | 2.021812 | CUUGGA,GUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| TRA2B | 30 | 7329 | 0.043055556 | 0.0106226650 | 2.019054 | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AGAAGA,AGAAGG,AGGAAA,AGGAAG,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| PABPC3 | 4 | 1234 | 0.006944444 | 0.0017897669 | 1.956088 | AAAAAC,AAAACA,AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPM | 3 | 999 | 0.005555556 | 0.0014492040 | 1.938671 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM41 | 4 | 1318 | 0.006944444 | 0.0019115000 | 1.861154 | AUACUU,UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 | 2 | 822 | 0.004166667 | 0.0011926949 | 1.804669 | UGUAGG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| AGO1 | 1 | 548 | 0.002777778 | 0.0007956130 | 1.803792 | GGUAGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| CSTF2 | 6 | 1967 | 0.009722222 | 0.0028520334 | 1.769295 | GUGUGU,GUGUUU,UGUGUG,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| PABPC4 | 3 | 1251 | 0.005555556 | 0.0018144033 | 1.614436 | AAAAAG | AAAAAA,AAAAAG |
| CELF2 | 5 | 1886 | 0.008333333 | 0.0027346479 | 1.607539 | GUCUGU,GUGUGU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ERI1 | 1 | 632 | 0.002777778 | 0.0009173461 | 1.598393 | UUCAGA | UUCAGA,UUUCAG |
| RBM24 | 6 | 2357 | 0.009722222 | 0.0034172229 | 1.508462 | AGUGUG,GUGUGG,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| TUT1 | 1 | 678 | 0.002777778 | 0.0009840095 | 1.497187 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| CELF1 | 6 | 2391 | 0.009722222 | 0.0034664959 | 1.487808 | GUGUGU,UGUCUG,UGUGUG,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| PABPC5 | 6 | 2400 | 0.009722222 | 0.0034795387 | 1.482390 | AGAAAA,AGAAAG,AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBMX | 13 | 4925 | 0.019444444 | 0.0071387787 | 1.445609 | AAGAAG,AAGGAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,GAAGGA,GGAAGG,GUAACA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ESRP2 | 5 | 2150 | 0.008333333 | 0.0031172377 | 1.418626 | GGGAAG,GGGGAG,GGGGAU,UGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBFOX1 | 2 | 1077 | 0.004166667 | 0.0015622419 | 1.415276 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| MATR3 | 1 | 739 | 0.002777778 | 0.0010724109 | 1.373073 | AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| NONO | 3 | 1498 | 0.005555556 | 0.0021723567 | 1.354670 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SART3 | 6 | 2634 | 0.009722222 | 0.0038186524 | 1.348223 | AAAAAC,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| TARDBP | 6 | 2654 | 0.009722222 | 0.0038476365 | 1.337314 | GAAUGG,GUGAAU,GUGUGU,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SRSF5 | 22 | 8869 | 0.031944444 | 0.0128544391 | 1.313298 | AACAGC,AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA,UACAGC,UGCAGA,UGCAGC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| MSI1 | 6 | 2770 | 0.009722222 | 0.0040157442 | 1.275619 | AGGAAG,AGUAAG,UAGGUG,UAGUAA,UAGUUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ZCRB1 | 3 | 1605 | 0.005555556 | 0.0023274215 | 1.255199 | GAAUUA,GAUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SNRNP70 | 2 | 1237 | 0.004166667 | 0.0017941145 | 1.215622 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| DAZAP1 | 13 | 5964 | 0.019444444 | 0.0086445016 | 1.169503 | AGAUAU,AGGAAA,AGGAAG,AGUAAG,AGUAUG,AGUUUA,UAGUAA,UAGUAU,UAGUUA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ZNF638 | 3 | 1773 | 0.005555556 | 0.0025708878 | 1.111665 | GGUUGG,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SRSF10 | 29 | 13860 | 0.041666667 | 0.0200874160 | 1.052602 | AAAAGA,AAAGAA,AAAGGA,AAGAAA,AAGAAG,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGG,AGAGAA,AGAGGA,GAAAGA,GAGAAG,GAGGAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| PUM2 | 3 | 1890 | 0.005555556 | 0.0027404447 | 1.019521 | GUAAAU,UAGAUA,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
RBP on BSJ (Exon Only)
Plot
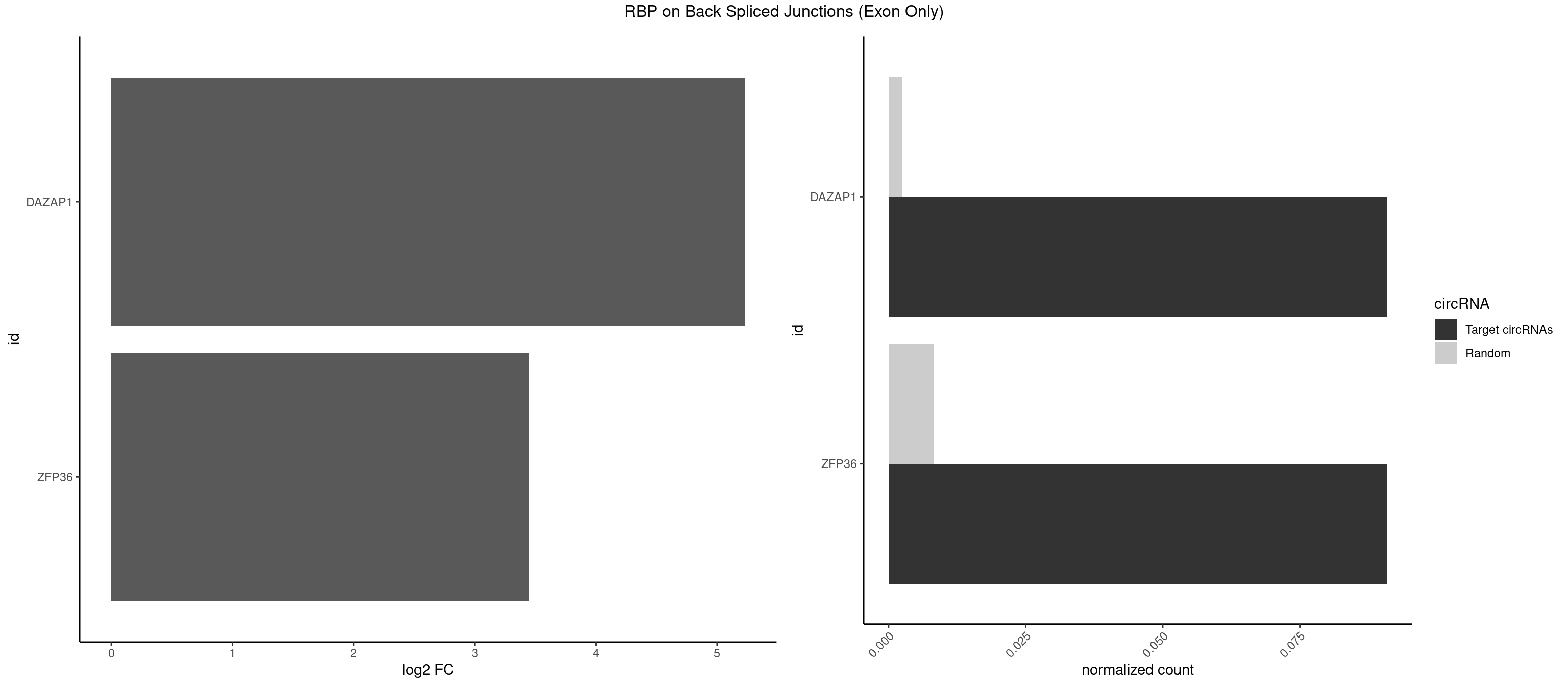
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| DAZAP1 | 1 | 36 | 0.09090909 | 0.002423369 | 5.229338 | AGUUUA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.008318051 | 3.450107 | AAAAGU | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
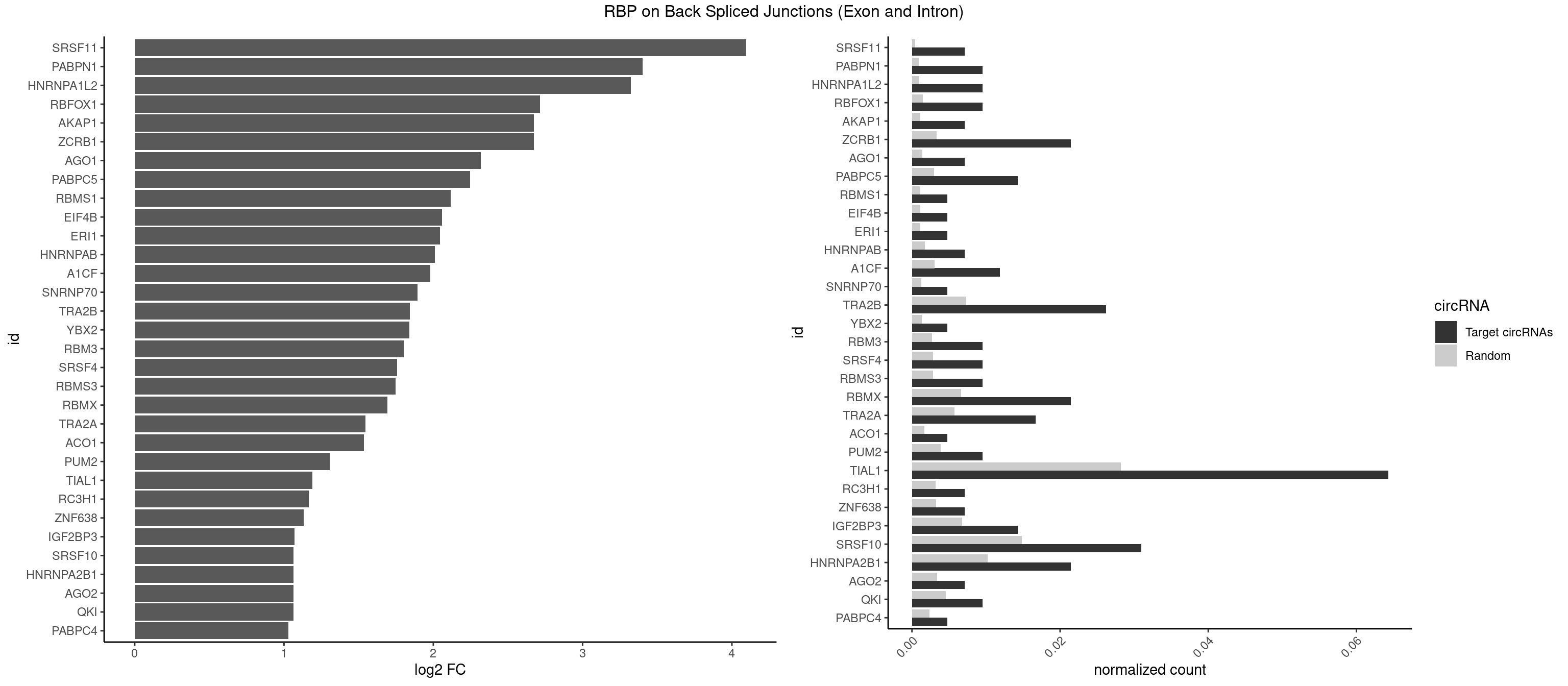
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF11 | 2 | 82 | 0.007142857 | 0.0004181782 | 4.094312 | AAGAAG | AAGAAG |
| PABPN1 | 3 | 178 | 0.009523810 | 0.0009018541 | 3.400573 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| HNRNPA1L2 | 3 | 188 | 0.009523810 | 0.0009522370 | 3.322146 | AUAGGG,GUAGGG,UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBFOX1 | 3 | 287 | 0.009523810 | 0.0014510278 | 2.714464 | AGCAUG,GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| ZCRB1 | 8 | 666 | 0.021428571 | 0.0033605401 | 2.672771 | ACUUAA,AUUUAA,GACUUA,GAUUUA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | GAGGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| PABPC5 | 5 | 596 | 0.014285714 | 0.0030078597 | 2.247764 | AGAAAA,AGAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AAAGAC,AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,AUCAGU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| TRA2B | 10 | 1447 | 0.026190476 | 0.0072954454 | 1.843974 | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AGAAGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AAACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SRSF4 | 3 | 558 | 0.009523810 | 0.0028164047 | 1.757684 | AAGAAG,AGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAU,CUAUAU,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBMX | 8 | 1316 | 0.021428571 | 0.0066354293 | 1.691274 | AAGAAG,AAGUAA,AAGUGU,AGUGUU,UAACAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| TRA2A | 6 | 1133 | 0.016666667 | 0.0057134220 | 1.544539 | AAAGAA,AAGAAA,AAGAAG,AGAAGA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UACAUC,UAUAUA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| TIAL1 | 26 | 5597 | 0.064285714 | 0.0282043531 | 1.188580 | AAAUUU,AAUUUU,AUUUUC,AUUUUG,AUUUUU,CUUUUA,CUUUUU,GUUUUU,UAAAUU,UAUUUU,UUAAAU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CCUUCU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUCUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAUUC,AAUACA,AAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPA2B1 | 8 | 2033 | 0.021428571 | 0.0102478839 | 1.064210 | AAGAAG,AUAGCA,AUAGGG,AUUUAA,GUAGGG,UAGACU,UAGGGU | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| SRSF10 | 12 | 2937 | 0.030952381 | 0.0148024990 | 1.064210 | AAAAGA,AAAGAA,AAAGAC,AAGAAA,AAGAAG,AAGAGA,AGAGAA,GAGAAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | AUCUAC,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.